ABSTRACT
The poultry red mite (PRM), Dermanyssus gallinae (De Geer, 1778), is a worldwide distributed ectoparasite and considered a major pest affecting the laying hen industry in Europe. Based on available information in other ectoparasites, the mite microbiome might participate in several biological processes and the acquisition, maintenance and transmission of pathogens. However, little is known about the role of PRM as a mechanical carrier or a biological vector in the transmission of pathogenic bacteria. Herein, we used a metaproteomics approach to characterize the alphaproteobacteria in the microbiota of PRM, and variations in its profile with ectoparasite development (nymphs vs. adults) and feeding (unfed vs. fed). The results showed that the bacterial community associated with D. gallinae was mainly composed of environmental and commensal bacteria. Putative symbiotic bacteria of the genera Wolbachia, C. Tokpelaia and Sphingomonas were identified, together with potential pathogenic bacteria of the genera Inquilinus, Neorickettsia and Roseomonas. Significant differences in the composition of alphaproteobacterial microbiota were associated with mite development and feeding, suggesting that bacteria have functional implications in metabolic pathways associated with blood feeding. These results support the use of metaproteomics for the characterization of alphaproteobacteria associated with the D. gallinae microbiota that could provide relevant information for the understanding of mite-host interactions and the development of potential control interventions.
Research highlights
Metaproteomics is a valid approach for microbiome characterization in ectoparasites.
Alphaproteobacteria putative bacterial symbionts were identified in D. gallinae.
Mite development and feeding were related to variations in bacterial community.
Potentially pathogenic bacteria were identified in mite microbiota.
Introduction
The haematophagous ectoparasite Dermanyssus gallinae (De Geer, 1778; Acari: Mesostigmata), commonly known as the poultry red mite (PRM), is the major pest of the poultry industry especially affecting laying hens (Chauve, Citation1998). D. gallinae s. str. is a complex generalist species with a low host specificity (Roy et al., Citation2009a, Citation2009b), and is becoming problematic due to its capacity to parasitize a wide range of domestic and wild birds, and even mammals including humans (Cafiero et al., Citation2008; Valiente Moro et al., Citation2009a; George et al., Citation2015). Interestingly, recent reports in pigeons have identified a lineage different from those in poultry, D. gallinae lineage 1 (Pezzi et al., Citation2017). D. gallinae requires a blood meal for moulting from protonymph to deutonymph, from deutonymph to adult and for egg-laying (Kilpinen, Citation2001). In laying hens, PRM is associated with poor health status and welfare problems associated with weight gain, reduced egg laying, and anaemia causing death in severe cases of infestation (Cosoroaba, Citation2001).
To date, the role of D. gallinae as a biological vector is questionable. Although its capacity as a reservoir of bacteria and viruses multiplying in the mite under field conditions has not been fully elucidated (Valiente Moro et al., Citation2005), the multiplication and transovarial/transstadial transmission of Salmonella enteritidis have been demonstrated (Valiente Moro et al., Citation2007). Several reports have identified pathogenic bacteria such as Coxiella burnetii, Erysipelothrix rhusiopathiae, Listeria monocytogenes, Pasterella multocida and Spirochetes in PRM (Valiente Moro et al., Citation2009b; Huong et al., Citation2014), demonstrating its role as mechanical vector for several pathogens (Sikes & Chamberlain, Citation1955; Zemskaya & Pchelkina, Citation1967; Ciolca et al., Citation1968; Shirinov et al., Citation1972; Petrov, Citation1975; Durden et al., Citation1993; Sommer et al., Citation2016). The presence of endosymbiotic bacteria, such as Spiroplasma, Candidatus Cardinium, Schineria, Ricketsiella, and Wolbachia spp., has been also described in D. gallinae (De Luna et al., Citation2009; Valiente Moro et al., Citation2009b; Hubert et al., Citation2017). The relationships between these bacterial symbionts and PRM has not been fully characterized, but it will be interesting to explore them as potential mechanisms for the control of PRM (De Luna et al., Citation2009).
Recent advances in metagenomics and metaproteomics have greatly contributed to the knowledge of the complexity and diversity of the ectoparasite microbiome, and its functional implications (Carpi et al., Citation2011; Neelakanta & Sultana, Citation2013; Chandler et al., Citation2015; Hubert et al., Citation2017; Swei & Kwan, Citation2017; Greay et al., Citation2018; Hernández-Jarguin et al., Citation2018). Hubert et al. (Citation2017) were the first to characterize the microbiome of farm-collected PRM using 16S ribosomal RNA metagenomics.
In this study, we used a metaproteomics approach focused on the class alphaproteobacteria because it is a wide diverse and abundant proteobacterial group within arthropods that includes medically important vector-borne pathogenic bacteria (i.e: the genera Anaplasma, Rickettsia, Brucella) and other important symbiotic genera (i.e: Wolbachia, or symbiont-like Sphingomonas) (Walker, Citation2017). In this study, we used a metaproteomic approach to identify microbial communities harboured by PRM. To date, metagenomics tools have been applied to unravel the composition of the microbial communities of mites (Hubert et al., Citation2017). The innovative aspect of this study was to complement current metagenomics information at protein levels and provide new insights into potential functional implications of mite microbiota.
Materials and methods
Mite collection and protein extraction
D. gallinae were collected from a free-range poultry unit in North East England. Mites were stored at 4°C within sealable plastic bags in complete darkness until used. Mites were sorted into groups: (i) engorged female adult mites (FA), (ii) non-fed female adult mites (UA), (iii) engorged proto and deutonymphs (FN), and (iv) unfed proto and deutonymphs (UN). The 1.5 ml tubes containing approximately 0.05 g of mites per sample were snap-frozen in liquid nitrogen and stored at −80°C until used for protein extraction. The mites were resuspended in ice-cold PBS (in a proportion of 10 ml per gram of mites) supplemented with cOmpleteTM Protease Inhibitor Cocktail (Roche Diagnostics GmbH, Mannheim, Germany), and homogenized on ice for two pulses of 30 s each with Ultra Turrex® T 25 D-S2 with a S25N-8G dispersing element (IKA, Sataufen, Germany). After centrifugation at 5000 × g for 20 min at 4°C, insoluble material and debris were removed and soluble material was decanted and centrifuged for a second time. The resulting soluble proteins were immediately snap-frozen and stored at −80°C until used for proteomics analysis.
Proteomics data acquisition and analysis
Protein concentration was determined using the BCA Protein Assay (Thermo Scientific, San Jose, CA, USA) with bovine serum albumin as standard. Protein extracts (75 µg per sample) were on-gel concentrated by SDS-PAGE as previously described (Villar et al., Citation2014). The unseparated protein bands were visualized by GelCode Blue Stain Reagent (Thermo Scientific, Waltham, MA, USA) excised, cut into 2 × 2 mm cubes and digested overnight at 37°C with 60 ng/μl sequencing grade trypsin (Promega, Madison, WI, USA) at 5:1 protein:trypsin (w/w) ratio in 50 mM ammonium bicarbonate, pH 8.8 containing 10% (v/v) acetonitrile (Shevchenko et al., Citation2007). The resulting tryptic peptides were extracted by incubation with 12 mM ammonium bicarbonate, pH 8.8, and digestion was stopped by the addition of trifluoroacetic acid to a final concentration of 1%. Peptides were finally desalted onto OMIX Pipette tips C18 (Agilent Technologies, Santa Clara, CA, USA), dried-down and stored at −20°C until mass spectrometry analysis.
The desalted protein digests were resuspended in 0.1% formic acid and analysed by reverse phase liquid chromatography coupled to mass spectrometry (RP-LC-MS/MS) using an Easy-nLC II system coupled to an LTQ-Orbitrap-Velos-Pro mass spectrometer (Thermo Scientific). The peptides were concentrated on-line by reverse phase chromatography using a 0.1 × 20 mm C18 RP precolumn (Thermo Scientific, Rockford, IL, USA) and then separated using a 0.075 × 250 mm C18 RP column (Thermo Scientific) operating at 300 nl/min. Peptides were eluted using a 140-min gradient from 5 to 40% solvent B in solvent A (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile). Electrospray ionization (ESI) was done using a Nano-bore Stainless Steel emitter ID 30 µm (Thermo Scientific) interface. Peptides were detected in survey scans from 400 to 1600 amu (1 µscan), followed by 20 data-dependent MS/MS scans (Top 20), using an isolation width of two mass-to-charge ratio units, normalized collision energy of 35%, and dynamic exclusion applied during 30 s periods.
The MS/MS raw files were searched against a compiled database containing the Uniprot alphaproteobacteria, parasitiformes and Gallus gallus databases (11,040,380, 141,928 and 29,484 entries in April 2018, respectively) together with a database created from the predicted secretome and transmembranome of D. gallinae (Schicht et al., Citation2013) using the SEQUEST algorithm (Proteome Discoverer 1.4, Thermo Scientific). The following constraints were used for the searches: tryptic cleavage after Arg and Lys, two maximum missed cleavages, and tolerances of 20 ppm for precursor ions and 0.05 Da for MS/MS fragment ions. The searches were performed allowing variable Met oxidation and Cys carbamidomethylation modifications. Searches were also performed against a decoy database in an integrated decoy approach. Three biological replicates per sample were analysed. A false discovery rate <0.01 was considered as the condition for successful peptide assignments, and at least two peptides per protein in the average of the replicates per sample (including at least one proteotypic peptide) were the necessary condition for protein identification. After discarding host and mite protein assignations, peptides corresponding to the taxonomic class alphaproteobacteria were grouped by genera taxa, and matches that did not reach the genera definition were discarded. The total number of peptide spectrum matches (PSMs) for each bacterial genus was normalized against the total number of PSMs to calculate taxonomic relative abundance at genus level. Raw proteomics data are available through the PeptideAtlas repository (http://www.peptideatlas.org/) with the dataset identifier PASS01346.
Alphaproteobacteria taxonomic assignments and phenotype visualization
A phylogenetic pruned tree associated to a heatmap was constructed to visualize the relative abundance of the peptide assignments (PSMs) corresponding to alphaproteobacteria identifications at genus level. The heatmap and pruned phylogenetic tree were generated with the platform PhyloT (https://phylot.biobyte.de) based on NCBI taxonomy, and visualized using the Interactive Tree of Life software v4.3 (https://itol.embl.de/) (Letunic and Bork, Citation2011). The alphaproteobacterial taxonomy-to-phenotype mapping was done using the platform METAGENassist (http://www.metagenassist.ca/METAGENassist/faces/Home.js) (Arndt et al., Citation2012). Furthermore, metabolic interpretations of the taxonomy-to-phenotype identifications for each bacteria genera were confirmed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (https://www.genome.jp/kegg/pathway.html).
Statistical analysis
The significance of metaproteomics comparative analysis between developmental and feeding stages was calculated by comparing the average amount of PSMs per bacterial genera for each group using a paired comparison Chi2-test (P < 0.05) and R software package (R Core Team, Citation2016). Additionally, bacterial genera distribution across the developmental stages and feeding status was visualized based on the Detrended Correspondence Analysis included in the vegan package in R software (Oksanen et al., Citation2018).
Results and discussion
Metaproteomics identification of alphaproteobacteria in the microbiota of D. gallinae
The metaproteomics analysis of the PRM resulted in a total of 2837 PSMs that matched with specific bacterial genera present in the alphaproteobacteria database resulting in the identification of 122 bacterial proteins distributed among 53 genera of the alphaproteobacteria (Supplementary Data 1).
The most represented genus found in the PRM metaproteome was Sphingomonas (58 PSMs) followed by Bradyrhizobium (51 PSMs), Rhodopseudomonas (49 PSMs), Methylobacterium (40 PSMs), and Wolbachia spp. (38 PSMs). As expected, most of the bacterial genera identified by metaproteomics have been previously described in different arthropods including PRM (Supplementary Table 1). In terms of taxonomic relative abundance, 74% (39 out of 53) of the identified genera have been previously reported as environmental bacteria (Salter et al., Citation2014; Razzauti et al., Citation2015; Degli Esposti & Martinez Romero, Citation2017; Hernández-Jarguín et al., Citation2018) (Supplementary Table 1). From these environmental bacteria, 20% (8 out of 39) have been identified in several mite species including D. gallinae, and 44% (17 out of 39) have also been found in arthropods other than mites, and in humans (Supplementary Table 1). These genera are common members of the soil and water microbial communities that might colonize bird breeding sites, and have been described as mite surface contamination (Hubert et al., Citation2017). Additionally, the genus Sphingomonas is commonly found in the microbiota of arthropods such as mites and ticks, suggesting a symbiotic role for these bacteria (Hubert et al., Citation2015; Gurfield et al., Citation2017; Hernández-Jarguín et al., Citation2018). Wolbachia spp. have also been described in different arthropods with biological relevance in mites associated to cytoplasmic incompatibility in phytophagus mites (Breeuwer & Jacobs, Citation1996; Breeuwer, Citation1997), and fecundity success in predatory mites (Weeks & Stouthamer, Citation2004).
Bacteria of the Caulobacter, Ensifer, Mesorhizobium, Rhizobium, Phillobacterium, Sphingopyxis and Wolbachia genera were identified in PRM by both metagenomics (Hubert et al., Citation2017) and metaproteomics approaches (, ). However, Bartonella-like bacteria were previously identified by metagenomics as core microbiota members of D. gallinae (Hubert et al., Citation2017); nevertheless this genera was not identified herein by metaproteomics. It has been discussed previously that a metaomics approach combining multiple omics technologies provides a better characterization of the tick bacterial microbiome by increasing bacterial identification and support for identified bacteria with putative functional implications (Hernández-Jarguín et al., Citation2018). However, metaproteomics may result in some peptide assignments that could result in matches that do not reach the genera definition and are therefore discarded for further analysis, a limitation that requires further analyses with amino acid sequences of peptides used for protein identity (Tanca et al., Citation2014; Fernández de Mera et al., Citation2017).
Figure 1. Phylogenetic and taxonomic abundance analyses at different feeding status and developmental stages. Phylogenetic pruned tree and associated heatmap showing the corresponding peptide assignments (PSMs) at genus level comparing fed (red coloured) to unfed (white coloured) adults and nymphs (Feeding chart) and fed adults vs. fed nymphs (red coloured) and unfed adults vs. unfed nymphs (white coloured) (Development chart). The analysis was done using a Heatmap Tool associated with a pruned phylogenetic tree generated with the platform PhyloT (http://phylot.biobyte.de) based on NCBI taxonomy, and visualized using the Interactive Tree of Life software v3.4.3 (http://itol.embl.de). Colour online.
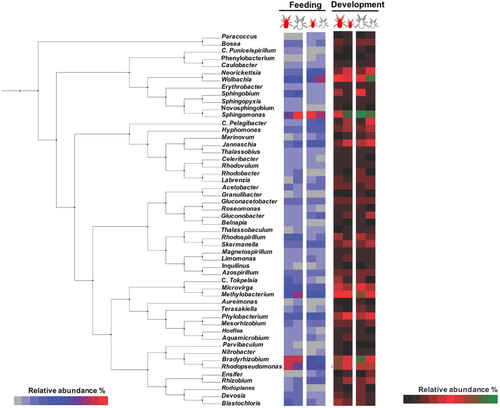
Table 1. Metaproteomics bacterial assignment at genus level and comparative analysis.
Differences in the profile of alphaproteobacteria in the microbiota associated with development and feeding of D. gallinae
Differences in bacterial microbiota composition between developmental stages and feeding status were observed (, , Supplementary Figure 1). These results are in accordance with previous reports in blood-feeding arthropods (Carpi et al., Citation2011; Hubert et al., Citation2017).
After differential analysis by comparing PSMs matching each genus, Candidatus Tokpelaia were significantly over-represented in fed adults when compared to fed nymphs (P = 0.034) and Sphingobium spp. were over-represented in fed and unfed adults when compared to fed and unfed nymphs (P = 0.035 and P = 0.025, respectively) (). The genus C. Tokpelaia has been found within the gut microbiota of ants and mites with an attributed symbiotic role participating in the urea cycle (Neuvonen et al., Citation2016; Hubert et al., Citation2018). Therefore, over-representation of bacteria from this genus in fed adults could be associated with the nitrogen and sulfur metabolism that occurs within the urea cycle after blood feeding (). In the same way, bacteria within the genus Sphingobium participate in the carbon fixation pathway (Zhang et al., Citation2012) (). Altogether, these results indicate that bacteria in these two genera are differentially represented in response to mite development and feeding and, in turn, could be implicated in facilitating blood digestion in adult mites using sulfur and carbon as energy sources ().
Figure 2. Identified bacterial microbiota phenotype. The bacterial taxonomy-to-phenotype mapping was done using the platform METAGENassist (http://www.metagenassist.ca/METAGENassist/faces/Home.js). Heatmaps were generated with genera classified using PSMs relative abundance. The dendrogram was generated using the similarity measure distance Pearson's correlation, and clustering using Ward's linkage algorithm. The heatmap displays an increase and decrease in each metabolic function in terms of genus PSMs relative abundance. Highlighted genera marked with an asterisk showed significant differences in pairwise comparisons (Chi2-test, P < 0.05) fully disclosed in .
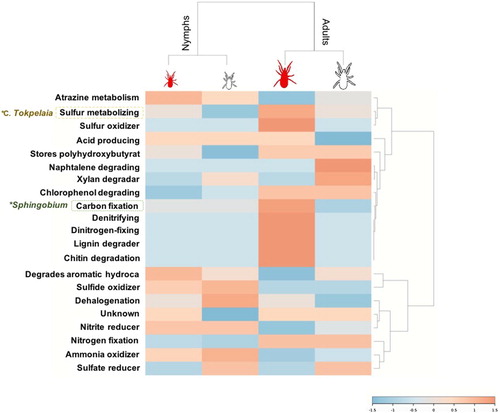
Potentially pathogenic alphaproteobacteria were identified in the microbiota of D. gallinae
Other bacterial genera identified in the metaproteome of PRM include some potentially pathogenic species for birds and humans, including Neorickettsia, Inquilinus, and Roseomonas spp. (Dumler et al., Citation2001; Coenye et al., Citation2002; Wellinghausen et al., Citation2005; Degli Esposti & Martinez Romero, Citation2017) (, , Supplementary Figure 1). Most of these bacteria have been reported in the microbial communities of blood-feeding arthropods, including ticks (Liu et al., Citation2010; Degli Esposti & Martinez Romero, Citation2017; Hernández-Jarguín et al., Citation2018). However, their role as vectors for these bacteria has been only partially characterized in ticks (Hernández-Jarguín et al., Citation2018).
Conclusion
The results of this study support the use of metaproteomics for the characterization of alphaproteobacteria associated with the microbiota of D. gallinae. Metaproteomics could be used in combination with other methods for a better identification and functional characterization of bacterial genera by a metaomics approach. The results of these studies could provide information relevant for the understanding of mite-host interactions and the development of potential control interventions.
Supplemental Material
Download MS Word (35.6 KB)Acknowledgements
We thank Dra. Lourdes Mateos (UMR BIPAR, INRA, France) and Dra. Marinela Contreras (IREC, Spain) for laboratory support, and Roxana Triguero (IREC, Spain) for computational support. Sabiotec S. A. did not participate in this research.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
José Francisco Lima-Barbero http://orcid.org/0000-0002-2694-0215
Sandra Díaz-Sanchez http://orcid.org/0000-0002-7205-3174
Olivier Sparagano http://orcid.org/0000-0003-3141-310X
Robert D. Finn http://orcid.org/0000-0003-0002-9711
José de la Fuente http://orcid.org/0000-0001-7383-9649
Margarita Villar http://orcid.org/0000-0003-4172-9079
Additional information
Funding
References
- Arndt, D., Xia, J., Liu, Y., Zhou, Y., Guo, A.C., Cruz, J.A., Sinelnikov, I., Budwill, K., Nesbø, C.L. & Wishart, D.S. (2012). METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Research, 40, W88–W95. doi: 10.1093/nar/gks497
- Breeuwer, J.A.J. & Jacobs, G. (1996). Wolbachia: intracellular manipulators of mite reproduction. Experimental & Applied Acarology, 20, 421–434. doi: 10.1007/BF00053306
- Breeuwer, J.A. (1997). Wolbachia and cytoplasmic incompatibility in the spider mites Tetranychus urticae and T. turkestani. Heredity, 79, 41–47. doi: 10.1038/hdy.1997.121
- Cafiero, M., Camarda, A., Circella, E., Santagada, G., Schino, G. & Lomuto, M. (2008). Pseudoscabies caused by Dermanyssus gallinae in Italian city dwellers: a new setting for an old dermatitis. Journal of the European Academy of Dermatology and Venereology, 22, 1382–1383. doi: 10.1111/j.1468-3083.2008.02645.x
- Carpi, G., Cagnacci, F., Wittekindt, N.E., Zhao, F., Qi, J., Tomsho, L.P., Drautz, D.I., Rizzoli, A. & Schuster, S.C. (2011). Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One, 6, e25604. doi: 10.1371/journal.pone.0025604
- Chandler, J.A., Liu, R.M. & Bennett, S.N. (2015). RNA shotgun metagenomic sequencing of northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Frontiers in Microbiology, 06, 185. doi: 10.3389/fmicb.2015.00185
- Chauve, C. (1998). The poultry red mite Dermanyssus gallinae (De Geer, 1778): current situation and future prospects for control. Veterinary Parasitology, 79, 239–245. doi: 10.1016/S0304-4017(98)00167-8
- Ciolca, A., Tanase, I. & May, I. (1968). Role of the poultry red mite, Dermanyssus gallinae, in the transmission of spirochaetosis. Archivum Veterinarium Polonicum, 5, 207–215.
- Coenye, T., Goris, J., Spilker, T., Vandamme, P. & LiPuma, J.J. (2002). Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. Journal of Clinical Microbiology, 40, 2062–2069. doi: 10.1128/JCM.40.6.2062-2069.2002
- Cosoroaba, I. (2001). Massive Dermanyssus gallinae invasion in battery-husbandry raised fowls. Revue de Medecine Veterinaire (France), 152, 89–96.
- De Luna, C.J., Moro, C.V., Guy, J.H., Zenner, L. & Sparagano, O.A.E. (2009). Endosymbiotic bacteria living inside the poultry red mite (Dermanyssus gallinae). Experimental and Applied Acarology, 48, 105–113. doi: 10.1007/s10493-008-9230-2
- Degli Esposti, M. & Martinez Romero, E. (2017). The functional microbiome of arthropods. PLoS One, 12, e0176573. doi: 10.1371/journal.pone.0176573
- Dumler, J.S., Barbet, A.F., Bekker, C., Dasch, G.A., Palmer, G.H., Ray, S.C., Rikihisa, Y. & Rurangirwa, F.R. (2001). Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. International Journal of Systematic and Evolutionary Microbiology, 51, 2145–2165. doi: 10.1099/00207713-51-6-2145
- Durden, L.A., Linthicum, K.J. & Monath, T.P. (1993). Laboratory transmission of Eastern Equine Encephalomyelitis virus to chickens by chicken mites (Acari: Dermanyssidae). Journal of Medical Entomology, 30, 281–285. doi: 10.1093/jmedent/30.1.281
- Fernández de Mera, I.G., Chaligiannis, I., Hernández-Jarguín, A., Villar, M., Mateos-Hernández, L., Papa, A., Sotiraki, S., Ruiz-Fons, F., Cabezas-Cruz, A., Gortázar, C. & de la Fuente, J. (2017). Combination of RT-PCR and proteomics for the identification of Crimean-Congo hemorrhagic fever virus in ticks. Heliyon, 3, e00353. doi: 10.1016/j.heliyon.2017.e00353
- George, D.R., Finn, R.D., Graham, K.M., Mul, M.F., Maurer, V., Moro, C.V. & Sparagano, O.A. (2015). Should the poultry red mite Dermanyssus gallinae be of wider concern for veterinary and medical science? Parasites & Vectors, 8, 178. doi: 10.1186/s13071-015-0768-7
- Greay, T.L., Gofton, A.W., Paparini, A., Ryan, U.M., Oskam, C.L. & Irwin, P.J. (2018). Recent insights into the tick microbiome gained through next-generation sequencing. Parasites & Vectors, 11, 12. doi: 10.1186/s13071-017-2550-5
- Gurfield, N., Grewal, S., Cua, L.S., Torres, P.J. & Kelley, S.T. (2017). Endosymbiont interference and microbial diversity of the Pacific coast tick, Dermacentor occidentalis, in San Diego County, California. Peer Journal, 13, e3202. doi: 10.7717/peerj.3202
- Hernández-Jarguín, A., Díaz-Sánchez, S., Villar, M. & de la Fuente, J. (2018). Integrated metatranscriptomics and metaproteomics for the characterization of bacterial microbiota in unfed Ixodes ricinus. Ticks and Tick-Borne Diseases, 9, 1241–1251. doi: 10.1016/j.ttbdis.2018.04.020
- Hubert, J., Erban, T., Kamler, M., Kopecky, J., Nesvorna, M., Hejdankova, S., Titera, D., Tyl, J. & Zurek, L. (2015). Bacteria detected in the honeybee parasitic mite Varroa destructor collected from beehive winter debris. Journal of Applied Microbiology, 119, 640–654. doi: 10.1111/jam.12899
- Hubert, J., Erban, T., Kopecky, J., Sopko, B., Nesvorna, M., Lichovnikova, M., Schicht, S., Strube, C. & Sparagano, O. (2017). Comparison of microbiomes between red poultry mite populations (Dermanyssus gallinae): predominance of Bartonella-like bacteria. Microbial Ecology, 74, 947–960. doi: 10.1007/s00248-017-0993-z
- Hubert, J., Nesvorna, M., Sopko, B., Smrz, J., Klimov, P. & Erban, T. (2018). Two populations of mites (Tyrophagus putrescentiae) differ in response to feeding on feces-containing diets. Frontiers in Microbiology, 9, 2590. doi: 10.3389/fmicb.2018.02590
- Huong, C.T.T., Murano, T., Uno, Y., Usui, T. & Yamaguchi, T. (2014). Molecular detection of avian pathogens in poultry red mite (Dermanyssus gallinae) collected in chicken farms. Journal of Veterinary Medical Science, 76, 1583–1587. doi: 10.1292/jvms.14-0253
- Kilpinen, O. (2001). Activation of the poultry red mite, Dermanyssus gallinae (Acari: Dermanyssidae), by increasing temperatures. Experimental & Applied Acarology, 25, 859–867. doi: 10.1023/A:1020409221348
- Letunic, I. & Bork, P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research, 39, W475–W478. doi: 10.1093/nar/gkr201
- Liu, W., Zhang, F., Qiu, E.C., Yang, J., Xin, Z.T., Wu, X.M., Tang, F., Yang, H. & Cao, W.C. (2010). Roseomonas sp. isolated from ticks, China. Emerging Infectious Diseases, 16, 1177–1178. doi: 10.3201/eid1607.090166
- Neelakanta, G. & Sultana, H. (2013). The use of metagenomic approaches to analyze changes in microbial communities. Microbiology Insights, 6, 37–48. doi: 10.4137/MBI.S10819
- Neuvonen, M.M., Tamarit, D., Näslund, K., Liebig, J., Feldhaar, H., Moran, N.A., Guy, L. & Andersson, S.G. (2016). The genome of Rhizobiales bacteria in predatory ants reveals urease gene functions but no genes for nitrogen fixation. Scientific Reports, 6, 39197. doi: 10.1038/srep39197
- Oksanen, J., Blanchet, F.G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P.R., ÓHara, R.B., Simpson, G.L., Solymos, P., Stevens, M.H.H., Szoecs, E. & Wagner, H. (2018). vegan: Community Ecology Package. R package version 2.5-2 https://CRAN.R-project.org/package=vegan.
- Petrov, D. (1975). Study of Dermanyssus gallinae as a carrier of Pasteurella multocida. Veterinarno-Meditsinski Nauki, 12, 32–36.
- Pezzi, M., Leis, M., Chicca, M. & Roy, L. (2017). Gamasoidosis caused by the special lineage L1 of Dermanysuss gallinae (Acarina: Dermanyssidae): a case of heavy infestation in a public place in Italy. Parasitology International, 66, 666–670. doi: 10.1016/j.parint.2017.05.001
- R Core Team. (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. http://www.R-project.org.
- Razzauti, M., Galan, M., Bernard, M., Maman, S., Klopp, C., Charbonnel, N., Vayssier-Taussat, V., Eloit, M. & Cosson, J.F. (2015). A comparison between transcriptome sequencing and 16S metagenomics for detection of bacterial pathogens in wildlife. PLoS Neglected Tropical Diseases, 9, e0003929. doi: 10.1371/journal.pntd.0003929
- Roy, L., Dowling, A.P.G., Chauve, C.M. & Buronfosse, T. (2009a). Delimiting species boundaries within Dermanyssus Duges, 1834 (Acari: Dermanyssidae) using a total evidence approach. Molecular Phylogenetics and Evolution, 50, 446–470. doi: 10.1016/j.ympev.2008.11.012
- Roy, L., Dowling, A.P.G., Chauve, C.M., Lesna, I., Sabelis, M.W. & Buronfosse, T. (2009b). Molecular phylogenetic assessment of host range in five Dermanyssus species. Experimental and Applied Acarology, 48, 115–142. doi: 10.1007/s10493-008-9231-1
- Salter, S.J., Cox, M.J., Turek, E.M., Calus, S.T., Cookson, W.O., Moffatt, M.F., Turner, P., Parkhill, J., Loman, N.J. & Walker, A.W. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology, 12, 87. doi: 10.1186/s12915-014-0087-z
- Schicht, S., Qi, W., Poveda, L. & Strube, C. (2013). The predicted secretome and transmembranome of the poultry red mite Dermanyssus gallinae. Parasites & Vectors, 6, 259. doi: 10.1186/1756-3305-6-259
- Shevchenko, A., Tomas, H., Havli, J., Olsen, J.V. & Mann, M. (2007). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols, 1, 2856–2860. doi: 10.1038/nprot.2006.468
- Shirinov, F.B., Ibragimova, A.I. & Misirov, Z.G. (1972). Spread of fowl pox virus by the mite Dermanyssus gallinae. Veterinariya (Moscow), 4, 48–49.
- Sikes, R.K. & Chamberlain, R.W. (1955). Laboratory Investigations on the role of bird mites in the transmission of eastern and western equine encephalitis 1. The American Journal of Tropical Medicine and Hygiene, 4, 106–118. doi: 10.4269/ajtmh.1955.4.106
- Sommer, D., Heffels-Redmann, U., Köhler, K., Lierz, M. & Kaleta, E.F. (2016). Role of the poultry red mite (Dermanyssus gallinae) in the transmission of avian influenza A virus. Tierärztliche Praxis Großtiere, 1, 47–54.
- Swei, A. & Kwan, J.Y. (2017). Tick microbiome and pathogen acquisition altered by host blood meal. The ISME Journal, 11, 813–816. doi: 10.1038/ismej.2016.152
- Tanca, A., Palomba, A., Pisanu, S., Deligios, M., Fraumene, C., Manghina, V., Pagnozzi, D., Addis, M.F. & Uzzau, S. (2014). A straightforward and efficient analytical pipeline for metaproteome characterization. Microbiome, 2, 49. doi: 10.1186/s40168-014-0049-2
- Valiente Moro, C., Chauve, C. & & Zenner, L. (2005). Vectorial role of some dermanyssoid mites (Acari, Mesostigmata, Dermanyssoidea). Parasite, 12, 99–109. doi: 10.1051/parasite/2005122099
- Valiente Moro, C., Chauve, C. & & Zenner, L. (2007). Experimental infection of Salmonella enteritidis by the poultry red mite, Dermanyssus gallinae. Veterinary Parasitology, 146, 329–336. doi: 10.1016/j.vetpar.2007.02.024
- Valiente Moro, C., De Luna, C.J., Tod, A., Guy, J.H., Sparagano, O.A.E. & Zenner, L. (2009a). The poultry red mite (Dermanyssus gallinae): a potential vector of pathogenic agents. Experimental and Applied Acarology, 48, 93–104. doi: 10.1007/s10493-009-9248-0
- Valiente Moro, C., Thioulouse, J., Chauve, C., Normand, P. & & Zenner, L. (2009b). Bacterial taxa associated with the hematophagous mite Dermanyssus gallinae detected by 16S rRNA PCR amplification and TTGE fingerprinting. Research in Microbiology, 160, 63–70. doi: 10.1016/j.resmic.2008.10.006
- Villar, M., Popara, M., Mangold, A.J. & de la Fuente, J. (2014). Comparative proteomics for the characterization of the most relevant Amblyomma tick species as vectors of zoonotic pathogens worldwide. Journal of Proteomics, 105, 204–216. doi: 10.1016/j.jprot.2013.12.016
- Walker, D.H. (2017). Rickettsia. In International Encyclopedia of Public Health (pp. 370–377). Second Edition. Quah: Academic Press.
- Weeks, A.R. & Stouthamer, R. (2004). Increased fecundity associated with infection by a Cytophaga–like intracellular bacterium in the predatory mite, Metaseiulus occidentalis. Proceedings of the Royal Society of London. Series B: Biological Sciences, 271 Suppl 4:S193-5.
- Wellinghausen, N., Essig, A. & Sommerburg, O. (2005). Inquilinus limosus in patients with cystic fibrosis, Germany. Emerging Infectious Diseases, 11, 457–459. doi: 10.3201/eid1103.041078
- Zemskaya, A. & Pchelkina, A. (1967). Gamasoid mites and Q fever. In Markevich (Ed.), Problemy Parazitologii (pp. 289–259). Kiev.
- Zhang, J., Lang, Z.F., Zheng, J.W., Hang, B.J., Duan, X.Q., He, J. & Li, S.P. (2012). Sphingobium jiangsuense sp. nov., a 3-phenoxybenzoic acid-degrading bacterium isolated from a wastewater treatment system. International Journal of Systematic and Evolutionary Microbiology, 62, 800–805. doi: 10.1099/ijs.0.029827-0
