ABSTRACT
Infectious bursal disease (IBD) is one of the most important immunosuppressive diseases of young chickens, causing considerable economic losses to the poultry industry. More than 30 years ago, an antigenic variant (av) pathotype of the IBD virus (IBDV) was reported to originate in, and subsequently spread among, poultry farms in the USA. Recently, a novel avIBDV lineage was identified in China and was shown to exhibit clear differences in its pathogenicity as well as molecular characteristics compared with the previously isolated variant strains. In this study, we conducted a passive surveillance of chicken carcasses submitted to our research division from June–December 2019, and detected the IBDV strains by reverse transcription PCR. Five avIBDV strains were isolated, and their pathogenicity was determined by necropsy and molecular analysis. Additionally, a coinfection field case involving an avIBDV strain and a very virulent IBDV (vvIBDV) strain was identified. Multiple sequence alignment and phylogenetic analysis of partial viral protein 1 (VP1) and hypervariable region (hv) VP2 genes revealed that those strains originated from two different avIBDV lineages. The co-occurrence of two sub-groups of avIBDVs in South Korea confirms for the first time the evolution of antigenic variant IBDV strains, and highlights the urgency for the development of new strategies for IBDV intervention in South Korea.
RESEARCH HIGHLIGHTS
Five avIBDV strains were identified in South Korea by passive surveillance test in 2019.
A coinfection between two IBDV strains from different genogroups was reported in a field case.
By phylogenetic analysis, Korean avIBDVs belonged to two distinct lineages of antigenic variant genogroup.
Introduction
Infectious bursal disease (IBD), commonly known as Gumboro disease, is an acute and highly contagious immunosuppressive viral disease of young chickens and is caused by the IBD virus (IBDV), which belongs to the Avibirnavirus genus of the Birnaviridae family. IBDV attacks and destroys B-lymphocytes in the bursa of Fabricius of young chickens, thus inducing immunosuppression and consequently high susceptibility to other diseases (Kibenge et al., Citation1988). IBDV has a non-enveloped capsid containing a double-stranded RNA genome with two segments, A and B. Segment A contains two overlapping open reading frames that encode viral proteins (VPs) including VP5 and VP2–VP4–VP3 (Muller et al., Citation2003). Previous studies mainly focused on the hypervariable region of VP2 (hvVP2) between amino acid (aa) positions 206 and 350 since it contains four hydrophilic regions determining virulence and antigenic variation (Bayliss et al., Citation1990). Segment B encodes VP1, an RNA-dependent RNA polymerase, which plays an important role in viral replication. Both VP1 and VP2 contribute to the virulence of IBDV and have been used to study its pathogenicity, phylogenetic relationship, and molecular characteristics (Boot et al., Citation2005; Brandt et al., Citation2001).
Two serotypes (I and II) of IBDV have been identified; however, only serotype I is pathogenic to chickens. Serotype I was originally classified into four main pathotypes, depending on their pathogenicity and antigenicity: classical virulent (cv), antigenic variant (av), very virulent (vv), attenuated (at) (van den Berg et al., Citation2004). However, due to rapid genetic variation in hvVP2 region, new classification of IBDVs with seven genogroups (G) has been proposed. According to this classification, cv/atIBDV, avIBDV and vvIBV strains were identified as G1, G2 and G3, respectively. G4 contained a group of virus strains that included the distinct (d) IBDV strains identified in the South American study which have spread rapidly through many regions around the world including Korea (Hernández et al., Citation2015; Kwon et al., Citation2000). G5 contained viruses isolated in Mexico and known to have genetic recombination between variant and classical viruses (Jackwood, Citation2012). Viruses in G6 were most closely related to the dIBDV from Italy (Lupini et al., Citation2016) and G7 contained viruses isolated in Australia (Michel & Jackwood, Citation2017). Immunisation of broiler chicks with intermediate or intermediate-plus strains and immunisation of breeders with live attenuated/inactivated vaccines is widely used to control IBDV infection worldwide. However, IBD outbreaks in vaccinated flocks have been documented in several countries (Geerligs et al., Citation2015). To date, vvIBDV strains have received the most attention because these strains cause acute infection, with high mortality and significant clinical signs. However, avIBDV infection is considered subclinical, and a diagnostic approach in the field is irrelevant. This genogroup is mostly prevalent in poultry farms in North America and Europe and further geographically distributed into three sub-lineages G2a, G2b, G2c with three representative strains Variant E, T1 and 3_Ohio, respectively (Lasher & Davis, Citation1997; Jackwood et al., Citation2006; Jackwood et al., Citation2018). Whereas only a few studies reported the occurrence of avIBDV in Asia until 2019, a large number of avIBDVs, identified as novel aetiological pathogens, were reported in China, causing widespread infection in chicken farms (Fan et al., Citation2019). In South Korea, the first IBD outbreak was reported in 1980 and four genogroups of IBDV (G1-G4) have been isolated to date, of which only one antigenic variant strain of G2 was isolated in 2010 (Jeon et al., Citation2009; Kim et al., Citation2010; Kwon et al., Citation2000). IBD in South Korea is mainly caused by vvIBDV and is controlled by vaccines (both live and killed) developed from cvIBDV. Given the recent prevalence of avIBDV in Asia, determining whether this genogroup has spread to Korea is important. In this study, we conducted a passive surveillance of 2–9-week-old chicken carcasses and isolated avIBDVs. Additionally, we performed a molecular analysis of VP1 and VP2 of five novel avIBDV strains identified in South Korea.
Materials and methods
Sample preparation
Carcasses from 2-9-week-old chickens, submitted to the Avian Diseases Research Division of Animal and Plant Quarantine Agency (APQA) from June to December 2019, were tested to detect and isolate IBDV. Bursas were homogenized in 10% (w/v) phosphate buffered saline (PBS) supplemented with 50 µg/ml gentamycin. The homogenate was centrifuged at 3000 × g for 10 min at 4°C, and the supernatant was collected for subsequent IBDV detection and analysis.
RNA extraction and reverse transcription PCR (RT–PCR)
Viral RNA was extracted from bursa homogenates using a Viral Gene-Spin DNA/RNA Extraction Kit (Intron Biotechnology, Seongnam, South Korea). Full-length VP1 and VP2 sequences were amplified from IBDV-infected samples using the primers listed in (Felice et al., Citation2017; Kong et al., Citation2004; Michel & Jackwood, Citation2017) RT–PCR was performed using Maxime RT–PCR Premix (Intron Biotechnology) under the following thermocycling conditions: 48°C for 30 min; 94°C for 10 min; 35 cycles of denaturation at 94°C for 30 s, annealing under primer-specific conditions, and extension at 72°C for 1.5 min; and final extension at 72°C for 10 min.
Table 1. List of primers used to amplify full-length IBDV viral protein 1 (VP1) and VP2 sequences.
Sequencing and phylogenetic analysis
The RT–PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany), and both cDNA strands were sequenced by Sanger sequencing using PCR primers (Bionics, Seoul, South Korea). Full-length nucleotide and amino acid sequences of VP1 and VP2 were edited and assembled using the CLC Workbench version 6.7.2 (CLC bio, Aarhus, Denmark). A 360-bp fragment (661–1021 nt) of hvVP2 and a 1051-bp fragment (319–1369 nt) of VP1 were aligned, and phylogenetic trees were constructed using the neighbor-joining method and 1000 bootstrap replications with the MEGA 7 software (Kumar et al., Citation2016). Full-length or partial coding sequences of VP1 and VP2 genes of five newly identified avIBDV strains were submitted to the GenBank database under the accession numbers MT550875–MT550883.
Virus isolation
In certain cases, bursa homogenate was inoculated into 11-day-old specific pathogen-free (SPF) embryonated chicken eggs via the chorioallantoic membrane (CAM) route. At 5 days post-inoculation, the CAM was homogenized, and RNA was extracted and analysed. Isolated IBDV strains were then screened for negative detection of adventitious pathogens including reovirus, adenovirus, fowl pox virus, or other viruses that also attack lymphoid organs, such as chicken anaemia virus (Saif & Jackwood, Citation2016).
Cloning
The hypervariable region of the VP2 gene was amplified and cloned into the TOPO TA vector. The resulting vector was transformed into HIT-DH5α competent cells using the TOPO TA Cloning Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. Three independent positive clones were sequenced, and the assembled sequences were analysed further.
Histological examination
Bursal tissues were fixed in 10% neutral-buffered formalin for histopathologic examination. Paraffin-embedded tissues were routinely processed, and 4 µm thick sections were prepared and stained with haematoxylin and eosin (H&E).
Results
Detection and diagnostic case report of avIBDVs
Of the 29 carcasses under passive surveillance, nine were positive for IBDV, and five of these nine cases (17.1% of all examined cases) carried avIBDV.
Details of the chickens carrying these five avIBDV strains, based on consultation with farm owners and field veterinarians, are summarized in . Chickens originated from four broiler farms and one Korean native chicken farm. All infected farms reported to have low mortality but carried cases of infectious bronchitis and bacterial infection. Necropsy revealed that five out of six examined chickens infected with the strain 19D75 contained an enlarged bursa of Fabricius with haemorrhage. No obvious gross lesion was detected in the bursas of chicken carcasses infected with other strains. Histopathological analysis revealed lymphocyte depletion in the bursa follicles of chickens infected with any of the five strains. Blood congestion was detected in the bursa of 19D38 ((a,b)). Development of cystic cavities in the bursa follicles was evident in 19D51 and 19D69. Loss of epithelium was observed in H&E-stained bursa sections of chickens infected with 19D51 and 19D69 strains, and lymphocytolysis and heterophil infiltration was detected with infection by the latter strain ((c,d)). Severe histopathological lesions were observed in bursa of 19D75, including follicle destruction ((e)) along with massive lymphocyte reduction, epithelium loss, karyorrhectic debris, and degenerated granulocytes ((f)).
Figure 1. Representative histological lesions in bursa sections of chickens infected with Korean antigenic variant infectious bursal disease virus (avIBDV) strains isolated in this study. (a, b) Bursa of 19D38-infected chickens showed mild lymphoid reduction and congestion. (c, d) Bursa follicles of chickens infected with 19D69 were infiltrated with mononuclear inflammatory cells, and showed lymphoid depletion, loss of follicular epithelium, cysts, and connective tissue formation. (e, f) Severe bursal lesions caused by 19D75 including follicle destruction, massive lymphocyte depletion, and granulocyte degeneration. Tissues were subjected to H&E staining, and images were captured at 40× magnification (a, c, e) or 100× magnification (b, d, f).
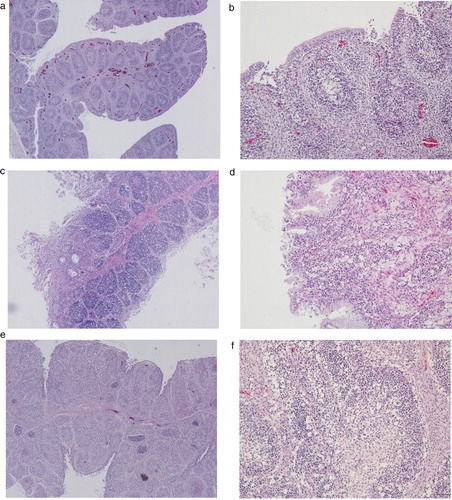
Table 2. Summary of Korean variant strains of IBDV identified in 2019 via passive surveillance of 2–9-week-old chicken carcasses.
Molecular characterization of Korean avIBDVs
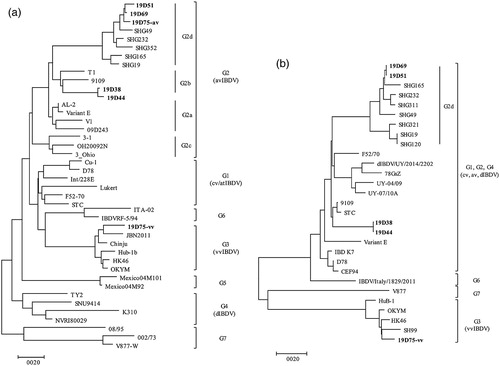
Phylogenetic analysis of the hvVP2 nucleotide sequences of serotype I strains revealed that these strains were divided into seven major branches according to seven genogroups ((a)). The antigenic variant group (G2) was further divided into four sub-lineages, one of which contained novel strains that recently originated from China. According to the phylogenetic analysis, five Korean IBDV strains identified in this study belonged to G2 major branch. While 19D38 and 19D44 were clustered with the representative avIBDV strains from G2b lineage which were demonstrated in America (T1, and 9109 strains), 19D51, 19D69, and 19D75 clustered together with SHG49, SHG19 and SHG232 which recently emerged in China, and clearly belonged to a new variant sub-lineage named G2d. Phylogenetic analysis of the VP1 nucleotide sequence showed that the serotype I was divided into four major clades: genogroup 3, genogroup 6, genogroup 7 and the remaining genogroups including G1, G2 and G4 ((b)). Although Korean avIBDVs were close to strains from genogroup 1 and 2, 19D38 and 19D44 formed an independent branch. Additionally, 19D51, 19D69, and novel Chinese variants formed a sub-branch, distinct from other genogroups.
Figure 2. Phylogenetic analysis of the nucleotide sequences of viral protein 2 (VP2) and VP1. Phylogenetic trees of hypervariable region of VP2 (a) and partial VP1 (b) generated by the neighbor-joining method using MEGA X with 1000 bootstrap replication. Korean IBDV strains identified in this study are indicated in bold text.
Further molecular characterization of Korean avIBDVs was conducted by multiple alignments of the deduced amino acid sequences of hvVP2 and VP1 (). According to the hvVP2 amino acid sequence alignment, five Korean avIBDV strains possessed 222T, 242V, 256V, 279N, and 294L residues, which are highly conserved among the IBDV strains in genotype 2. However, 19D38 and 19D44 were more closely related to variant strains from G2a and G2b lineages (93.5–96.1% amino acid sequence identity) than to the novel variants G2d (91.5–92.9% sequence identity) which were considered as new sub-lineages. Additionally, 19D51, 19D69, and 19D75 showed 95.6–98.6% sequence similarity with the G2d, which was considerably higher than the similarity to the previous other lineages (91.7–94.0%). The 19D51, 19D69, and 19D75 strains also contained highly conserved amino acid residues in VP2 including 221K, 252I, and 299S, and in VP1 (508K) which were detected only in the novel avIBDV lineage.
Table 3. Key amino acid residues in the hypervariable region of VP2 showing phylogenic characteristics of Korean antigenic variant IBDVs and reference strains.
Initially, sequencing the hvVP2 of strain 19D75 amplified by RT–PCR showed a double peak at certain nucleotides, suggesting a possible coinfection of two different IBDV strains in a non-immunized flock. Attempts to isolate this virus were performed by virus inoculation into SPF embryonated eggs via the CAM. Notably, the partial nucleotide sequence of VP2 isolated from the bursa homogenate was closely related to the novel variant lineage, whereas the nucleotide sequence of the strain isolated by CAM was similar to vvIBDV. This result indicates a coinfection of 19D75 chickens with avIBDV and vvIBDV strains. Unfortunately, while the vvIBDV strain in 19D75 was successfully isolated, the avIBDV strain could not be isolated.
Discussion
The avIBDV strains originated in North America over 30 years ago, and became widespread in the USA and recently in Europe. VP2, the antigenic determinant of IBDV, is most commonly used for the molecular characterization of the virus. Given the high potency of mutations in the hypervariable region of VP2, Jackwood et al. (Citation2018) proposed a new universal naming system for IBDVs, according to which avIBDVs were identified as Genogroup 2 and geographically divided into three sub-lineages with American origin. In Korea, an antigenic variant strain 09D243 was reported in 2010 and it was close to avIBDV strain Variant E in the phylogenetic tree (Kim et al., Citation2010). Besides, a group of antigenic variant-like strains including K310, SNU9414 and NVRI80029 were isolated with a particular antigenic profile and named by the authors as Korean antigenic variant IBDVs (Jeon, Citation2009; Kwon, Citation2000). However, these strains were considered as distinct IBDVs and later classified as genogroup 4 (Tomás et al., Citation2019). In 2019, a major epidemic of avIBDV occurred in China, and this has now been identified in poultry farms in South Korea (Fan et al., Citation2019) According to the results of hvVP2 phylogenetic analysis and key amino acid comparison among seven IBDV genogroups, two Korean avIBDVs (19D51 and 19D69) identified in this study, along with novel avIBDV strains isolated in China, clearly belonged to G2. These strains made a new sub-branch within G2 and these strains have not been identified in any previous studies about IBDV nomenclature. Since Jackwood et al. (Citation2018) already classified three sub-lineages within G2, the novel variant group might be considered to be the new sub-lineage G2d. Defining new sub-lineages based on phylogenetic tree topology, in combination with biological characterization, has proven to be reasonably useful. Compared with the previously known G2 sub-lineages, strains in the novel variant lineage contained several amino acid substitutions, which could affect the antigenicity and pathogenicity of the strain as well as the host immune response. Therefore, it is critical to track the changes in avIBDV strains at the molecular level to gain a better understanding of the current epidemiological status of IBDV in Asia. Therefore, it is important to investigate the effect of these mutations in future studies.
Infectious bronchitis virus, as well as bacterial pathogens, was isolated in chickens infected with all five Korean variant IBDVs. Although we could not elucidate whether the in-field Korean avIBDV infection was primary or secondary, this coinfection is consistent with previous studies, suggesting that the main pathogenic property of avIBDVs may relate to immunosuppression, resulting in high susceptibility of IBDV-infected chickens to other infectious agents (Jackwood, Citation2012; Jackwood & Saif, Citation1987; Fan et al., Citation2020; Jackwood, Citation2012). Among the five Korean avIBDVs, three were isolated from chicken flocks immunized with the intermediate or intermediate-plus IBDV vaccines. However, necropsy and histological lesions revealed no differences between immunized and non-vaccinated chickens. This implies that the current virus management system in Korea could be invalidated by avIBDVs.
Bursal atrophy is one of the typical lesions found in avIBDV-infected chickens (Sharma et al., Citation1989; Xu et al., Citation2020). However, we hardly saw any atrophic signs in bursas collected from the field, except in those infected by 19D75, which was later identified as a coinfection field case. The avIBDVs could not have been recognized without the passive surveillance test. To some extent, histological lesions of two American-like avIBDVs, 19D38 and 19D44, were comparably less severe than those of 19D51 and 19D69, which belonged to the novel avIBDV lineage. Furthermore, strain 19D69 showed marcrophage infiltration in bursa follicles, which is consistent with a previous study, proving that novel variant IBDVs may exhibit relatively high pathogenicity (Fan et al., Citation2020; Xu et al., Citation2020). Accurate pathogenicity and immunosuppressive property of Korean avIBDVs in comparison with other sub-lineages of this genogroup should be carefully investigated to customize vaccine development in the future.
Different types of IBDVs were previously isolated from the same bursa, implying coinfection in the field (He et al., Citation2012). One pathogenicity study showed that coinfection between vvIBDV strain rB and avIBDV strains Del-E or T1 resulted in a significant reduction in mortality and severity of bursal lesions compared with infection by rB alone (Stoute et al., Citation2013). Furthermore, PCR and sequencing analysis detected only the rB strain, indicating a possible viral competition between vvIBDV and the less virulent pathotypes such as antigenic variants. It is also possible that the viral replicative property of vvIBDV is higher than that of other strains. Factors that influence the viral competition may include competition of cell surface receptors and intracellular mechanism required for viral replication. Host factors, such as nonspecific immunity or viral infection time, also act as possible determinants of which virus pathotype is dominant to the other (Jackwood, Citation2011). This finding is consistent with our histological examination of the bursas of 19D75 and with the successful isolation of only vvIBDV from the coinfection case.
In conclusion, here we report for the first time the co-occurrence of different sub-lineages of avIBDVs in South Korea. Passive surveillance revealed that 17% of the examined poultry farms were infected with avIBDVs. This information suggests that avIBDV may become endemic in Asia. Evidence showing that avIBDVs cannot be controlled using the current anti-IBDV vaccination strategy emphasize the urgency for further molecular analysis and pathogenicity tests for developing a new vaccine programme.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bayliss, C., Spies, U., Shaw, K., Peters, R., Papageorgiou, A., Müller, H. & Boursnell, M. (1990). A comparison of the sequences of segment A of four infectious bursal disease virus strains and identification of a variable region in VP2. Journal of General Virology, 71, 1303–1312.
- Boot, H.J., Hoekman, A.J. & Gielkens, A.L. (2005). The enhanced virulence of very virulent infectious bursal disease virus is partly determined by its B-segment. Archives of Virology, 150, 137–144.
- Brandt, M., Yao, K., Liu, M., Heckert, R.A. & Vakharia, V.N. (2001). Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. Journal of Virology, 75, 11974–11982.
- Fan, L., Wu, T., Hussain, A., Gao, Y., Zeng, X., Wang, Y., Gao, L., Li, K., Wang, Y., Liu, C., Cui, H., Pan, Q., Zhang, Y., Liu, Y., He, H., Wang, X. & Qi, X. (2019). Novel variant strains of infectious bursal disease virus isolated in China. Veterinary Microbiology, 230, 212–220.
- Fan, L., Wu, T., Wang, Y., Hussain, A., Jiang, N., Gao, L., Li, K., Gao, Y., Liu, C., Cui, H., Pan, Q., Zhang, Y., Wang, X. & Qi, X. (2020). Novel variants of infectious bursal disease virus can severely damage the bursa of Fabricius of immunized chickens. Veterinary Microbiology, 240, 108507.
- Felice, V., Franzo, G., Catelli, E., Di Francesco, A., Bonci, M., Cecchinato, M., Mescolini, G., Giovanardi, D., Pesente, P. & Lupini, C. (2017). Genome sequence analysis of a distinctive Italian infectious bursal disease virus. Poultry Science, 96, 4370–4377.
- Geerligs, H.J., Ons, E., Boelm, G.J. & Vancraeynest, D. (2015). Efficacy, safety, and interactions of a live infectious bursal disease virus vaccine for chickens based on strain IBDV877. Avian Diseases, 59, 114–121.
- He, X., Wei, P., Yang, X., Guan, D., Wang, G. & Qin, A. (2012). Molecular epidemiology of infectious bursal disease viruses isolated from Southern China during the years 2000-2010. Virus Genes, 45, 246–255.
- Hernández, M., Tomás, G., Marandino, A., Iraola, G., Maya, L., Mattion, N., Hernández, D., Villegas, P., Banda, A., Panzera, Y. & Pérez, R. (2015). Genetic characterization of South American infectious bursal disease virus reveals the existence of a distinct worldwide-spread genetic lineage. Avian Pathology, 44, 212–221.
- Jackwood, D.H. & Saif, Y.M. (1987). Antigenic diversity of infectious bursal disease viruses. Avian Diseases, 31, 766–770.
- Jackwood, D.J. (2011). Viral competition and maternal immunity influence the clinical disease caused by very virulent infectious bursal disease virus. Avian Diseases, 55, 398–406.
- Jackwood, D.J. (2012). Molecular epidemiologic evidence of homologous recombination in infectious bursal disease viruses. Avian Diseases, 56, 574–577.
- Jackwood, D.J., Cookson, K.C., Sommer-Wagner, S.E., Le Galludec, H. & de Wit, J.J. (2006). Molecular characteristics of infectious bursal disease viruses from asymptomatic broiler flocks in Europe. Avian Diseases, 50, 532–536.
- Jackwood, D.J., Schat, K.A., Michel, L.O. & de Wit, S. (2018). A proposed nomenclature for infectious bursal disease virus isolates. Avian Pathology, 47, 576–584.
- Jeon, W.J., Choi, K.S., Lee, D.W., Lee, E.K., Cha, S.H., Cho, S.H., Kwon, J.H., Yoon, Y.S., Kim, S.J., Kim, J.H. & Kwon, H.J. (2009). Molecular epizootiology of infectious bursal disease (IBD) in Korea. Virus Genes, 39, 342–351.
- Kibenge, F.S., Dhillon, A.S. & Russell, R.G. (1988). Biochemistry and immunology of infectious bursal disease virus. Journal of General Virology, 69, 1757–1775.
- Kim, H.R., Kwon, Y.K., Bae, Y.C., Oem, J.K. & Lee, O.S. (2010). Genetic characteristics of virion protein 2 genes of infectious bursal disease viruses isolated from commercial chickens with clinical disease in South Korea. Poultry Science, 89, 1642–1646.
- Kong, L.L., Omar, A.R., Hair-Bejo, M., Aini, I. & Seow, H.F. (2004). Sequence analysis of both genome segments of two very virulent infectious bursal disease virus field isolates with distinct pathogenicity. Archives of Virology, 149, 425–434.
- Kumar, S., Stecher, G. & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
- Kwon, H.M., Kim, D.K., Hahn, T.W., Han, J.H. & Jackwood, D.J. (2000). Sequence of precursor polyprotein gene (segment A) of infectious bursal disease viruses isolated in Korea. Avian Diseases, 44, 691–696.
- Lasher, H.N. & Davis, V.S. (1997). History of infectious bursal disease in the U.S.A. – the first two decades. Avian Diseases, 41, 11–19.
- Lupini, C., Giovanardi, D., Pesente, P., Bonci, M., Felice, V., Rossi, G., Morandini, E., Cecchinato, M. & Catelli, E. (2016). A molecular epidemiology study based on VP2 gene sequences reveals that a new genotype of infectious bursal disease virus is dominantly prevalent in Italy. Avian Pathology, 45, 458–464.
- Michel, L.O. & Jackwood, D.J. (2017). Classification of infectious bursal disease virus into genogroups. Archives of Virology, 162, 3661–3670.
- Muller, H., Islam, M.R. & Raue, R. (2003). Research on infectious bursal disease – the past, the present and the future. Veterinary Microbiology, 97, 153–165.
- Saif, Y.M. & Jackwood, D.J. (2016). Infectious bursal disease. In S.M. Williams, L. Dufour-Zavala, M.W. Jackwood, M.D. Lee, B. Lupiani, W.M. Reed, E. Spackman & P.R. Woolcock (Eds.), A Laboratory Manual for the Isolation, Identification, and Characterization of Avian Pathogens 6th edn (pp. 239–242). Athens, GA: American Association of Avian Pathologists.
- Sharma, J.M., Dohms, J.E. & Metz, A.L. (1989). Comparative pathogenesis of serotype 1 and variant serotype 1 isolates of infectious bursal disease virus and their effect on humoral and cellular immune competence of specific-pathogen-free chickens. Avian Diseases, 33, 112–124.
- Stoute, S.T., Jackwood, D.J., Sommer-Wagner, S.E., Crossley, B.M., Woolcock, P.R. & Charlton, B.R. (2013). Pathogenicity associated with coinfection with very virulent infectious bursal disease and infectious bursal disease virus strains endemic in the United States. Journal of Veterinary Diagnostic Investigation, 25, 352–358.
- Tomás, G., Marandino, A., Courtillon, C., Amelot, M., Keita, A., Pikula, A., Hernández, M., Hernández, D., Vagnozzi, A., Panzera, Y., Domańska-Blicharz, K., Eterradossi, N., Pérez, R. & Soubies, S.M. (2019). Antigenicity, pathogenicity and immunosuppressive effect caused by a South American isolate of infectious bursal disease virus belonging to the “distinct” genetic lineage. Avian Pathology, 48, 245–254.
- van den Berg, T.P., Morales, D., Eterradossi, N., Rivallan, G., Toquin, D., Raue, R., Zierenberg, K., Zhang, M.F., Zhu, Y.P., Wang, C.Q., Zheng, H.J., Wang, X., Chen, G.C., Lim, B.L. & Muller, H. (2004). Assessment of genetic, antigenic and pathotypic criteria for the characterization of IBDV strains. Avian Pathology, 33, 470–476.
- Xu, A., Pei, Y., Zhang, K., Xue, J., Ruan, S. & Zhang, G. (2020). Phylogenetic analyses and pathogenicity of a variant infectious bursal disease virus strain isolated in China. Virus Research, 276, 197833.
