ABSTRACT
Newcastle disease virus (NDV) affects commercial poultry as well as other avian species in the wild and in captivity. Although the diversity of NDV in domestic chickens has been well understood, little light has been shed on NDV outbreaks in other avian species. We provide an annotated sequence of NDV/Owl/Guwahati/01/20, a virulent strain of NDV isolated from barn owls in captivity from Guwahati in Northeast India. The complete genome is 15,192 base pairs long with a fusion protein (F) cleavage site 112KRQKR↓F117. The isolate showed 97.67% identity with its closest match, another highly virulent strain from Indonesia isolated from vaccinated commercial chickens; however, they differ in the F cleavage site. The NDV isolate from the owl shares 83.02% and 81.88% identity with the vaccine strains R2B and LaSota, respectively. Phylogenetic analysis with F gene as well as whole-genome nucleotide sequence reveals that the NDV isolate from owl belongs to genotype VII, subgenotype VII.2, and differs significantly from all other isolates of NDV from India.
Introduction
Newcastle disease virus (NDV), also known as Avian orthoavulavirus 1 is a century-old avian pathogen, first reported in 1926 (Kraneveld, Citation1926; Doyle, Citation1927). Ever since its first outbreak, NDV has been the cause of numerous pandemics in avian species, mostly among chickens (El-Nassary & Eskarous, Citation1960; Siccardi, Citation1966; Al-Hilly et al., Citation1980; Lomniczi et al., Citation1998). NDV is an enveloped negative-sense, single-stranded RNA virus having a genome length of 15,186, 15,192 or 15,198 nucleotides (Czegledi et al., Citation2006). The NDV genome codes for six open reading frames, namely nucleocapsid (N), phosphoprotein (P), matrix protein (M), fusion protein (F), haemagglutinin-neuraminidase (HN) and large polymerase protein (L) (Fields et al., Citation2013). The HN protein is responsible for the binding of virions to cellular sialic acid, a ubiquitous cellular receptor (Herrler et al., Citation1995). The F protein cleavage site determines the pathogenicity of NDV strains (Panda et al., Citation2004). NDV strains are broadly classified on the basis of virulence into lentogenic (least), mesogenic (moderately), and velogenic (highly) virulent (Alexander, Citation2001). Genetically, NDV is classified into class I and class II, which is further divided into 21 different genotypes (Dimitrov et al., Citation2019).
NDV continues to circulate among the avian population in captivity as well as in wild, leading to the emergence of new and diverse genetic variants (Jadhav et al., Citation2020). There have been numerous studies on NDV outbreaks in economically important domestic chickens (Awan et al., Citation1994).
However, little attention has been given to NDV isolated in other species and their diversity, which could create a possibility of unchecked NDV spread and the potential threat of an outbreak. Other than commercial chickens in poultry farms, exotic birds are generally kept in close proximity in zoos, making them vulnerable for spill-over from domestic birds leading to accidental outbreaks (Cardenas Garcia et al., Citation2013).
Materials and methods
Sample collection
Four barn owls (Tyto alba) were rescued in an adverse condition from a single nest at Sonaram HS School playground, Guwahati, India, after their nest was found broken. The birds were shivering and showing tremors as a neurological sign before death within 24 h. Tissue samples from lung, spleen, kidney, brain, and intestine were collected under aseptic conditions by the Office of the Forest Veterinary Officer, Assam State Zoo Division, Government of Assam, Guwahati. The samples were sent for diagnostic purposes to the Department of Microbiology, Assam Agricultural University (AAU), Khanapara, Guwahati, India, and for further molecular characterization to the Department of Biosciences and Bioengineering, IIT Guwahati, India.
Virus isolation and characterization
NDV was suspected to be the causative pathogen based on splenomegaly and haemorrhages in the intestines during the post-mortem examination. The presence of NDV was confirmed by PCR with NDV-specific primers using cDNA prepared from the lung tissue samples of infected birds. The tissue samples were triturated thoroughly with a mortar and pestle in phosphate buffered saline and centrifuged at 13,000 rpm for 10 min, 100 μl of supernatant along with 100 μl of 100 × antibiotic-antimycotic cocktail (Gibco, Thermo Fisher, USA) were inoculated into 9-day-old specific pathogen-free (SPF) embryonated chicken eggs. Allantoic fluid was harvested following the death of the embryos, and the presence of NDV was confirmed by haemagglutination (HA) assay using 1% chicken red blood cells. The virus was purified from the allantoic fluid through plaque purification in the chicken embryo fibroblast (CEF) cells (Khadzhiev, Citation1982). The isolated NDV was characterized for Mean Death Time (MDT) and Intracerebral Pathogenicity Index (ICPI) test following standard protocol in 9-day-old SPF embryonated chicken eggs and one-day-old SPF chicks, respectively (Terregino & Capua, Citation2009).
Genome sequence analysis
The Mean Death Time and Intracerebral Pathogenicity Index values of virus isolated from all the birds were found to be the same; one of them was selected for further analyses, including complete genome sequencing. The total RNA was isolated from the allantoic fluid using Trizol reagent (Takara, Shiga, Japan), and cDNA was synthesized using a Superscript III cDNA synthesis kit (Invitrogen, Waltham, Massachusetts, USA) following the manufacturer’s protocol. Furthermore, NDV-specific diagnostic primers available in the laboratory were used to confirm the genome (Stear, Citation2005). The primer sets spanning the entire genome (Supplementary Table 1) were used to amplify the viral genome in fragments using PCR with EmeraldAmp GoTaq Master Mix PCR kit (Takara, Japan). Each of the amplicons generated was sequenced with the same forward and reverse primer(s) using Sanger dideoxy sequencing technique at AgriGenome Labs Pvt. Ltd., Kochi, India. The fragment sequences spanning the whole NDV genome were then assembled to generate the whole genome using the EGassembler tool (Masoudi-Nejad et al., Citation2006). DNAStar bioinformatics tool was employed for translating the complete genome, retrieving protein sequences, pairwise and multiple sequence alignments, as well as for percentage identity calculations. The phylogenetic analysis was performed using the whole genome nucleotide sequence and only the F gene nucleotide sequence using the MEGA-X phylogenetic analysis tool (Kumar et al., Citation2018). Maximum likelihood trees were generated with 1000 bootstrap iterations.
Results and discussion
Virus characterization
Virus isolated from the spleen, brain, and intestines from all four owls were positive in HA, haemagglutination-inhibition, and PCR tests. The Mean Death Time was observed as ∼44.6 h, while an Intracerebral Pathogenicity Index value of 1.4375 was calculated for the NDV/Owl/Guwahati/01/20 isolate (hereafter abbreviated as NDV/Owl), suggesting the velogenic nature of the isolate. The 15,192 bp long whole genome of NDV/Owl was submitted to GenBank (accession number MZ546197).
Complete genome sequence analysis
The 15,192-bp long complete genome of NDV/Owl substantially differs from that of the vaccine strains R2B and LaSota, sharing 83.02% and 81.88% identity, respectively. The F protein of NDV/Owl showed 88.61% amino acid sequence identity, with both R2B and LaSota. The F cleavage site of NDV/Owl was found to be 112KRQKR↓F117(underlined letters represent basic residues, and the downward arrow represents the site of cleavage). The length of the HN protein of NDV/Owl was found to be 571 amino acids, which is a common feature of highly virulent strains of NDV (Zhao et al., Citation2013). The F protein of NDV/Owl was 555 amino acids long, two residues longer than most NDV isolates but similar to three unpublished isolates reported from Indonesia (GenBank accession numbers MW811474, MW811476, and MW811478). The phylogenetic analysis revealed that the NDV/Owl is significantly different from other previously reported NDV strains from India. The highly virulent Indonesian NDV isolate chicken/Banjarmasin/010/10 (Xiao et al., Citation2012) was found to be the closest match with NDV/Owl, with 97.67% and 98.73% identity in the complete genome and F protein amino acid sequence level, respectively. However, the F cleavage sites of NDV/Owl and chicken/Banjarmasin/010/10 (112RRQKR↓F117) were different. Phylogenetic analysis of NDV/Owl with the other NDV strains using whole genome and F gene suggested it to be of genotype VII (). Furthermore, subgenotyping of NDV/Owl following the recent classification showed that it belongs to subgenotype VII.2 (Dimitrov, et al., Citation2019) (). All genes of NDV/Owl except HN showed high identity with chicken/Banjarmasin/010/10. In contrast, the HN gene of NDV/Owl was found to share the highest identity of 99.12% with the isolate pigeon/Pakistan/Lahore/21A/1084/2015.
Figure 1. Phylogenetic analysis of NDV/Owl isolate based on F gene nucleotide sequence (a), and tree based on whole genome nucleotide sequence (b).
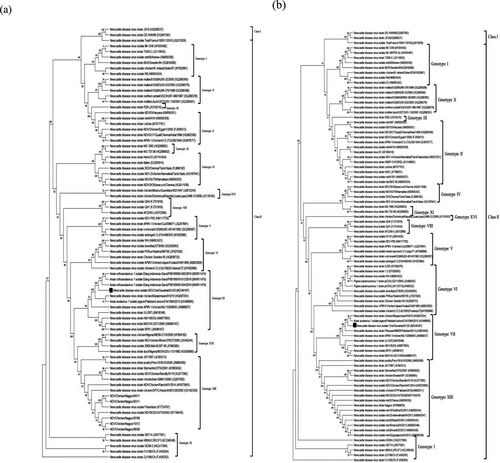
Figure 2. Phylogenetic analysis of NDV/Owl isolate. Phylogenetic tree based on F gene nucleotide sequence among genotype VII isolates (a), and phylogenetic tree based on whole genome nucleotide sequence among genotype VII isolates (b).
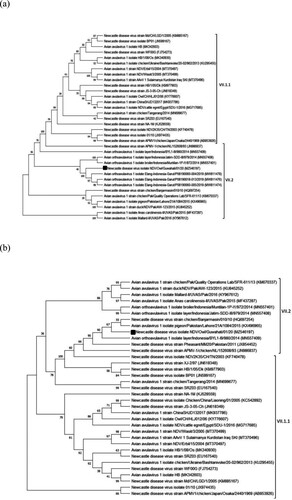
India is an NDV endemic country, and there have been many reports of NDV from the country in the recent past. Out of the 48 complete genome sequences of NDV reported from India, most isolates belong to genotype XIII. Although there have been some reports of genotypes I, II, and IV, NDV/Owl is only the second genotype VII complete genome from India, isolate NDV2K35/CH/TN/2003 (belonging to subgenotype VII.1.1), which was reported in 2003, being the first. In addition to having a different F cleavage site from that of NDV2K35/CH/TN/2003 (112RRQKR↓F117), NDV/Owl shares only 88.86% complete genome nucleotide identity and 95.66% F protein amino acid identity, respectively, with the former. This indicates that NDV/Owl is significantly different from NDV2K35/CH/TN/2003, and it is a unique NDV isolate in the Indian context.
The Mean Death Time and Intracerebral Pathogenicity Index values of NDV/Owl suggested that the strain is virulent. Further, the F cleavage site of isolate NDV/Owl 112KRQKR↓F117 confirmed its velogenic nature. The uniqueness of NDV/Owl and its high identity with some of the Indonesian isolates suggests an international spill-over. NDV is known to be spread across long geographical distances by migratory birds (Marks et al., Citation2014) and through unintentional human interventions (Chen & Wang, Citation2002). Many wild birds are also known to act as natural NDV reservoirs leading to the persistence of NDV in the wild and occasional spill-overs to domestic poultry (Brown & Bevins, Citation2017). Moreover, habitat sharing among domestic poultry with wild birds further aggravates the risk of introducing genetic variants (Shriner & Root, Citation2020). In addition, there are higher chances of infection in scavengers such as owls feeding on potentially infected bird carcasses (Reperant et al., Citation2008).
This study reports a virulent NDV isolate that differs significantly from any other isolate previously reported from India. As this isolate differs significantly from the R2B and LaSota strains, presumably the vaccines might not protect the birds in the event of a Newcastle disease outbreak, especially in endemic countries (Dimitrov et al., Citation2017). It is important to sequence more isolates of NDV from wild birds to understand the nature of circulating viruses. In addition, biosurveillance programmes for regular NDV testing, including real-time reverse transcriptase PCR and haemagglutination inhibition assay, should be introduced to tackle ND, especially in the endemic regions of the country (Zeynalova et al., Citation2015).
Ethical statement
All the bird experiments were approved by the Institutional Animal Care and Use Committee of AAU, Khanapara, Guwahati, India, and performed following the institutional guidelines.
Supplemental Material
Download MS Word (12.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Al-Hilly, J.N., Khalil, H.H., Zakoo, F.I. & Hamid, A.A. (1980). An outbreak of Newcastle disease in a pheasant flock in Iraq. Avian Pathology, 9, 583–585.
- Alexander, D.J. (2001). Gordon memorial lecture. Newcastle disease. British Poultry Science, 42, 5–22.
- Awan, M.A., Otte, M.J. & James, A.D. (1994). The epidemiology of Newcastle disease in rural poultry: a review. Avian Pathology, 23, 405–423.
- Brown, V.R. & Bevins, S.N. (2017). A review of virulent Newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Veterinary Research, 48, 68.
- Cardenas Garcia, S., Navarro Lopez, R., Morales, R., Olvera, M.A., Marquez, M.A., Merino, R., Miller, P.J., & Afonso, C.L. (2013). Molecular epidemiology of Newcastle disease in Mexico and the potential spillover of viruses from poultry into wild bird species. Applied and Environmental Microbiology, 79, 4985–4992.
- Chen, J.P. & Wang, C.H. (2002). Clinical epidemiologic and experimental evidence for the transmission of Newcastle disease virus through eggs. Avian Diseases, 46, 461–465.
- Czegledi, A., Ujvari, D., Somogyi, E., Wehmann, E., Werner, O. & Lomniczi, B. (2006). Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Research, 120, 36–48.
- Dimitrov, K.M., Abolnik, C., Afonso, C.L., Albina, E., Bahl, J., Berg, M., Briand, F.X., Brown, I.H., Choi, K.S., Chvala, I., Diel, D.G., Durr, P.A., Ferreira, H.L., Fusaro, A., Gil, P., Goujgoulova, G.V., Grund, C., Hicks, J.T., Joannis, T.M., Torchetti, M.K., Kolosov, S., Lambrecht, B., Lewis, N.S., Liu, H., McCullough, S., Miller, P.J., Monne, I., Muller, C.P., Munir, M., Reischak, D., Sabra, M., Samal, S.K., Servan de Almeida, R., Shittu, I., Snoeck, C.J., Suarez, D.L., Van Borm, S., Wang, Z. & Wong, F.Y.K. (2019). Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infection Genetics and Evolution, 74, 103917.
- Dimitrov, K.M., Afonso, C.L., Yu, Q. & Miller, P.J. (2017). Newcastle disease vaccines-A solved problem or a continuous challenge? Veterinary Microbiology, 206, 126–136.
- Doyle, T.M. (1927). A hitherto unrecognized disease of fowls due to a filter-passing virus. Journal of Comparative Pathology and Therapeutics, 40, 144–169.
- El-Nassary, B.B. & Eskarous, J.K. (1960). The distribution of fowl plague and Newcastle disease in upper Egypt. Archiv fuer Mikrobiologie, 36, 147–150.
- Fields, B.N., Knipe, D.M. & Howley, P.M. (2013). Fields Virology. Philadelphia, PA: Wolters Kluwer Health/Lippincott, Williams & Wilkins.
- Herrler, G., Hausmann, J. & Klenk, H.-D. (1995). Sialic acid as receptor determinant of ortho- and paramyxoviruses. In A. Rosenberg (Ed.), Biology of the Sialic Acids (315–336). Boston, MA: Springer US.
- Jadhav, A., Zhao, L., Liu, W., Ding, C., Nair, V., Ramos-Onsins, S.E. & Ferretti, L. (2020). Genomic diversity and evolution of quasispecies in Newcastle disease virus infections. Viruses, 12, 1305.
- Khadzhiev, G. (1982). Plaque formation by strains of Newcastle disease virus in monolayers of chick embryo fibroblasts. Veterinarno-Meditsinski Nauki, 19, 35–45.
- Kraneveld, F.C. (1926). Over een in Ned. Indie heerschende ziekte onder het pluimvee. Nederl.-Indische Bladen v. Diergeneesk, 38, 448–450.
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018). MEGA x: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549.
- Lomniczi, B., Wehmann, E., Herczeg, J., Ballagi-Pordany, A., Kaleta, E.F., Werner, O., Meulemans, G., Jorgensen, P.H., Mante, A.P., Gielkens, A.L., Capua, I. & Damoser, J. (1998). Newcastle disease outbreaks in recent years in Western Europe were caused by an old (VI) and a novel genotype (VII). Archives of Virology, 143, 49–64.
- Marks, F.S., Rodenbusch, C.R., Okino, C.H., Hein, H.E., Costa, E.F., Machado, G., Canal, C.W., Brentano, L. & Corbellini, L.G. (2014). Targeted survey of Newcastle disease virus in backyard poultry flocks located in wintering site for migratory birds from southern Brazil. Preventive Veterinary Medicine, 116, 197–202.
- Masoudi-Nejad, A., Tonomura, K., Kawashima, S., Moriya, Y., Suzuki, M., Itoh, M., Kanehisa, M., Endo, T. & Goto, S. (2006). EGassembler: online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Research, 34, W459–W462.
- Panda, A., Huang, Z., Elankumaran, S., Rockemann, D.D. & Samal, S.K. (2004). Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microbial Pathogenesis, 36, 1–10.
- Reperant, L.A., van Amerongen, G., van de Bildt, M.W., Rimmelzwaan, G.F., Dobson, A.P., Osterhaus, A.D. & Kuiken, T. (2008). Highly pathogenic avian influenza virus (H5N1) infection in red foxes fed infected bird carcasses. Emerging Infectious Diseases, 14, 1835–1841.
- Shriner, S.A. & Root, J.J. (2020). A review of avian influenza A virus associations in synanthropic birds. Viruses, 12, 1209.
- Siccardi, F.J. (1966). Effect of vaccination during an outbreak of Newcastle disease on a broiler-breeder chicken farm in Nigeria. Avian Diseases, 10, 422–427.
- Stear, M.J.J.P. (2005). OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds and Bees) 5th Edn. Volumes 1 & 2. Paris: World Organization for Animal Health 2004. 130, 727–727.
- Terregino, C. & Capua, I. (2009). Conventional diagnosis of Newcastle disease virus infection. In I. Capua & D.J. Alexander (Eds.), Avian Influenza and Newcastle Disease: A Field and Laboratory Manual (pp. 123–125). Milan: Springer.
- Xiao, S., Paldurai, A., Nayak, B., Samuel, A., Bharoto, E.E., Prajitno, T.Y., Collins, P.L. & Samal, S.K. (2012). Complete genome sequences of Newcastle disease virus strains circulating in chicken populations of Indonesia. Journal of Virology, 86, 5969–5970.
- Zeynalova, S., Guliyev, F., Vatani, M. & Abbasov, B. (2015). Biosurveillance of avian influenza and Newcastle disease viruses in the Barda region of Azerbaijan using real time RT-PCR and hemagglutination inhibition. Frontiers in Microbiology, 6, 1128.
- Zhao, W., Zhang, Z., Zsak, L. & Yu, Q. (2013). Effects of the HN gene C-terminal extensions on the Newcastle disease virus virulence. Virus Genes, 47, 498–504.
