ABSTRACT
Duck adenovirus 3 (DAdV-3) has been identified as the causative agent of a disease characterized by swelling and haemorrhages in liver and kidney of Muscovy ducks, causing huge economic losses to the waterfowl industry in China. In this study, a total of 54 DAdV-3 outbreaks from 2018 to 2020 in China were monitored with samples being collected and analyzed. The hexon amino acid sequences of the 54 DAdV-3 outbreaks and the DAdV-3 reference strains from GenBank were 98.7% to 100% identical. Epidemiological analysis showed that DAdV-3 circulated in meat-type and egg-laying Muscovy ducks, and co-infections with other viral and bacterial pathogens, such as Muscovy duck parvovirus (MDPV), Muscovy duck-origin goose parvovirus (MDGPV), adeno-associated virus (AAV), duck hepatitis B virus (DHBV), R. anatipestifer, and E. coli were identified. In addition, 12 out of the 23 (52.2%) DAdV-3 strains were isolated on LMH cells and identified by DAdV-3 real-time PCR. The whole-genome sequence analysis demonstrated that 12 new DAdV-3 isolates share 99.8–100% identity with the DAdV-3 reference strains, and nine new DAdV-3 isolates exhibit a truncated ORF67 gene. Our results enhance the understanding of the epidemiology and molecular characterization associated with DAdV-3 infection in China.
First report of the epidemiology of duck adenovirus 3 infection in China.
Mutant DAdV-3 strains (truncated ORF67) became predominant.
RESEARCH HIGHLIGHTS
Introduction
Adenoviruses (AdVs), belonging to the family Adenoviridae, are found in a wide variety of animals and humans. The Adenoviridae family is currently divided into five genera, including Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus and Ichtadenovirus, based on the International Committee on Taxonomy of Viruses (ICTV) (Harrach et al., Citation2011). Five adenoviruses originating from ducks have so far been identified. For example, duck adenovirus 1 (DAdV-1, genus Atadenovirus), reduces egg production and the quality of the eggs, and induces an acute respiratory disease (Cha et al., Citation2013). Duck adenovirus 2 (DAdV-2, genus Aviadenovirus) was detected from sick Muscovy ducks in France in 1977, and the complete genome of the DAdV-2 isolate GR was published in 2014 (Marek et al., Citation2014). Duck adenovirus 3 (DAdV-3, genus Aviadenovirus) was first isolated from diseased Muscovy ducks with haemorrhages and necrosis of the liver in China in 2014 (Zhang et al., Citation2016). Duck adenovirus 4 (DAdV-4, genus Aviadenovirus) was isolated from Jinding ducks with salpingitis in south China in 2019 (Huang et al., Citation2020). In addition, fowl adenovirus serotype 4 (FAdV-4, genus Aviadenovirus) infection associated with inclusion body hepatitis and hydropericardium syndrome has been reported in duck flocks in China (Chen et al., Citation2017; Pan et al., Citation2017; Yu et al., Citation2018).
DAdV-3 is a non-enveloped, double-stranded DNA virus with a genome approximately 44 kb in length, encoding some non-structural proteins and three main exposed structural proteins (penton base, hexon and fibre) (Zhang et al., Citation2016). The penton base plays a primary role in the virus attachment and endocytosis (Harrach et al., Citation2011; Harrach, Citation2014). The hexon protein, the most abundant viral surface protein, contains group-specific and type-specific antigenic determinants (Russell, Citation2009). The fibre protein, including a short fibre (fibre-1) and a long fibre (fibre-2), is responsible for cellular receptor binding and viral pathogenicity (Pallister et al., Citation1996; Russell, Citation2009).
In recent years, DAdV-3, a novel duck adenovirus, was reported as the aetiological agent of a disease characterized by swelling and haemorrhagic liver and kidney in Muscovy ducks, with a mortality rate up to 40%, leading to major economic losses in the poultry industry in China (Zhang et al., Citation2016; Shi et al., Citation2019). However, there have been no systematic epidemiological surveys of DAdV-3. In the present study, we collected tissue samples from Muscovy ducks showing gross lesions including swelling and haemorrhages of liver and kidney to gain an understanding of the epidemiology of DAdV-3 infection in China between 2018 and 2020.
Materials and methods
Sample collection and virus detection
Between January 2018 and December 2020, a total of 78 clinical cases suspected of DAdV-3 infection, characterized by gross lesions including swollen and haemorrhagic liver and kidney, in different commercial Muscovy duck farms in three provinces (including Guangdong, Fujian and Jiangxi) in China were selected for disease diagnosis. All tissue samples, such as livers and kidneys, were homogenized in phosphate buffered saline (PBS) to obtain a 10% tissue suspension. The suspensions were subjected to three freeze–thaw cycles before centrifugation at 5000 × g for 10 min. Then, the supernatant was used for virus isolation and virus nucleic acid extraction by using a MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa, Dalian, China), following the manufacturer’s protocols. The presence of DAdV-3 in samples was confirmed by real-time polymerase chain reaction (PCR), and the quantity of DAdV-3 DNA in tested samples was calculated based on the cycle threshold (Ct) values for the standard curve as previously described (Wan et al., Citation2018). The presence of other pathogens, including duck hepatitis virus types 1 and 3 (DHAV-1 and DHAV-3) (Zhang et al., Citation2017), duck hepatitis B virus (DHBV) (Wang et al., Citation2013), Muscovy duck parvovirus (MDPV) (Lin et al., Citation2019), Muscovy duck origin goose parvovirus (MDGPV) (Lin et al., Citation2019), Muscovy duck reovirus (MDRV) (Zheng et al., Citation2020), duck enteritis virus (DEV) (Hansen et al., Citation1999), adeno-associated virus (AAV) (Su et al., Citation2017), and FAdV-4 (Yu et al., Citation2018) were assessed by real-time PCR or conventional PCR using primers and methods as previously described. In addition, three possible bacterial pathogens, Riemerella anatipestifer (R. anatipestifer), Escherichia coli (E. coli), and Salmonella enterica (S. enterica), were isolated by the streak method on tryptic soy agar (TSA, Becton, Sparks, MD, USA) plates containing 5% foetal bovine serum at 37°C (Liu et al., Citation2018), and detected by multiplex PCR assay as previously described (Hu et al., Citation2011). In addition, liver and kidney samples were collected, fixed in 10% formalin, and stained with haematoxylin and eosin (H&E) for histopathological examination.
Virus isolation
Those tissue samples that were only positive for DAdV-3 were used to isolate DAdV-3 on Leghorn male hepatoma (LMH) cells (ATCC #CRL-2117). Briefly, LMH cell monolayers were cultured in 12-well plates, and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Logan, UT, USA) supplemented with 1% antibiotic-antimycotic (Gibco, Grand Island, NY, USA) and 10% foetal bovine serum (FBS) (BOVOGEN, Melbourne, Australia), and incubated at 37°C with 5% CO2. The DAdV-3 positive supernatant was filtered through a 0.22 µm syringe filter (Millipore, Tullagreen, Carrigtwohill, Ireland), and overlayed onto LMH cells. After 1 h absorption, the supernatant was discarded and replaced with fresh DMEM supplemented with 1% antibiotic-antimycotic and 2% FBS. After 72 h of incubation, the supernatant and cells were harvested by three freeze-thaw cycles for the next round of virus propagation and real-time PCR detection (Wan et al., Citation2018). The DAdV-3 isolate was obtained by the fifth passage in LMH cells and used for subsequent experiments. In addition, the cytopathic effect (CPE) was observed for 6 days, and the median tissue culture infectious dose (TCID50) was analyzed by the method of Reed & Muench (Citation1938).
Viral DNA extraction and PCR
The genomic DNA of DAdV-3 positive samples was extracted using a MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa), according to the manufacturer’s instructions. The hexon gene of the 54 DAdV-3 positive samples was amplified using a sense primer (5’-CAA TTT TAA ACG GTT TGT AGG TTC-3’) and an antisense primer (5’ -TAG CTG ACT GTC GGT GGT TCC T-3’) as previously described (Wan et al., Citation2018). The size of the amplified segment was about 2882 bp, and the complete hexon gene (2814 nt) was used for phylogenetic analysis. In addition, the whole genome of 12 DAdV-3 strains was amplified with 17 pairs of primers designed within conserved regions of the DAdV-3 sequences as previously described (Shi et al., Citation2019).
PCR was performed in a 50 μl volume containing 2 μl of DNA, 10 pmol of each gene-specific primer, 25 μl of PrimeSTAR Max Premix (2×) (TaKaRa). The thermal cycling parameters were as follows: 95°C for 5 min; 35 cycles of 98°C for 10 s, 55°C for 5 s, 72°C for 30 s; 72°C for 10 min.
Cloning and nucleotide sequencing
All PCR products were tested by electrophoresis in 1% agarose gels and purified by using the HiPure Gel Pure DNA Micro Kits (Magen, Guangzhou, China). All gel-purified PCR products were subjected to A-Tailing by using a DNA A-Tailing Kit (TaKaRa), and cloned into a pMD19-T vector (TaKaRa), and transformed into E. coli DH5α competent cells (TaKaRa). At least three positive clones of each PCR fragment were selected for DNA sequencing by GENEWIZ Company (Guangzhou, China).
Sequence comparison and phylogenetic analysis
The 17 overlapped nucleotide sequences obtained from the DAdV-3 strains were assembled into a complete genome sequence using the Seqman program of the DNAstar Lasergene software package (version 7.1) (DNAStar, Madison, WI, USA). Sequence alignments were performed using the Clustal W method, and phylogenetic trees of the full-length genome and hexon amino acid sequence were conducted by the neighbour-joining method with 1000 bootstrap replicates in the MEGA 7.0 software (Kumar et al., Citation2016).
Statistical analyses
All data were analyzed by using GraphPad Prism 6 software (version 6.01) (GraphPad Software, LaJolla, CA, USA).
Results
DAdV-3 epidemiology and pathology
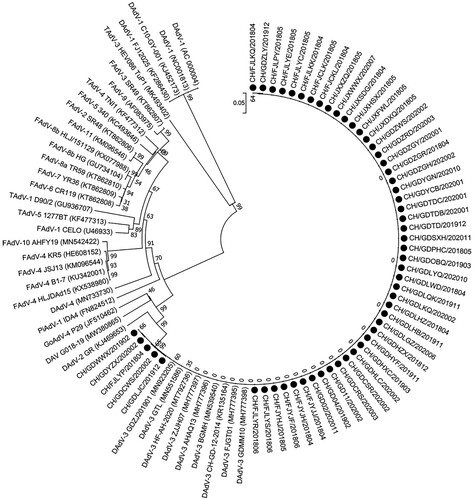
From 2018 to 2020, 78 clinical cases from Muscovy duck flocks suspected of a DAdV-3 infection, due to gross lesions in liver and kidney characterized by swelling and haemorrhagic, were examined. Of the 78 cases, 54 (69.23%) outbreaks tested positive for DAdV-3, and the epidemiological details of the 54 DAdV-3 positive outbreaks are described in Supplemental Table S1. The mortality rate ranged from 0.13% to 33.26%. Spatial analysis showed that DAdV-3 outbreaks appeared in three provinces, and 34 DAdV-3 outbreaks were detected in Guangdong province ((a)). Temporal analysis showed that the number of DAdV-3 outbreaks identified in 2018, 2019 and 2020 was 24, 10 and 20, respectively ((b)). Clinical cases of DAdV-3 infections occurred in meat-type Muscovy ducks that were 13–45 days old (average 24.1 days) and in layer Muscovy ducks which were 252–357 days old (average 308.0 days) ((c)). Interestingly, 66.7% (36 of 54 cases) of the DAdV-3 infections occurred in Muscovy ducks that were 25 days old or younger. In addition, we also screened for co-infections with other viral and bacterial pathogens, such as DHAV-1, DHAV-3, DHBV, MDPV, MDGPV, MDRV, DEV, FAdV-4, R. anatipestifer, E. coli and S. enterica. For these pathogens, 13 samples were positive for R. anatipestifer, six were positive for MDPV, four were positive for MDGPV, four were positive for AAV, two were positive for DHBV, two were positive for E. coli, and none was positive for FAdV-4 ((d)).
Figure 1. Summary of clinical cases (n = 54) of DAdV-3 infections in Muscovy ducks between 2018 and 2020 in China. (a) Geographical distribution of clinical cases of DAdV-3. (b) Temporal distribution of clinical cases of DAdV-3. (c) The age of the affected birds. (d) The presence of other viral and bacterial pathogens.
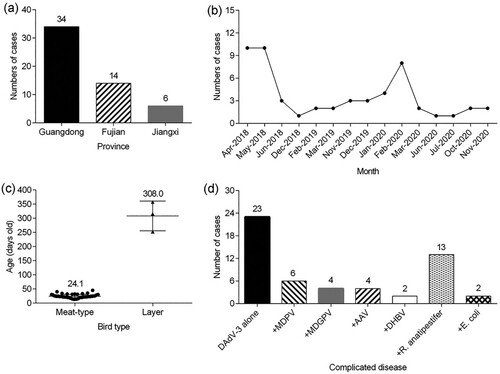
Most of the clinical cases caused by DAdV-3 infection alone were associated with a swollen and haemorrhagic liver ((a)); the same lesions were also noticed in kidneys ((b)). The most prominent microscopic lesions of DAdV-3 infection alone were degeneration of hepatocytes and inflammatory cell infiltration in the liver ((c)), as well as necrosis of renal tubules accompanied by local tubular epithelial cells detached from the basement membrane and interstitial congestion of the renal tubule in the kidney ((d)). No inclusion bodies were found in the liver and kidney.
Figure 2. Post mortem and histopathological analysis of tissues from dead Muscovy duck infected only with DAdV-3 in China. (a) Swelling, haemorrhage and necrosis in the liver (black arrow). (b) Enlarged kidney with haemorrhagic spots. (c) Degeneration of hepatocytes (black arrow) and inflammatory cells infiltration (white arrow) in the liver. (d) Necrosis of renal tubules accompanied by local tubular epithelial cells detached from the basement membrane (black arrow), interstitial congestion of the renal tubule (white arrow) in the kidney.
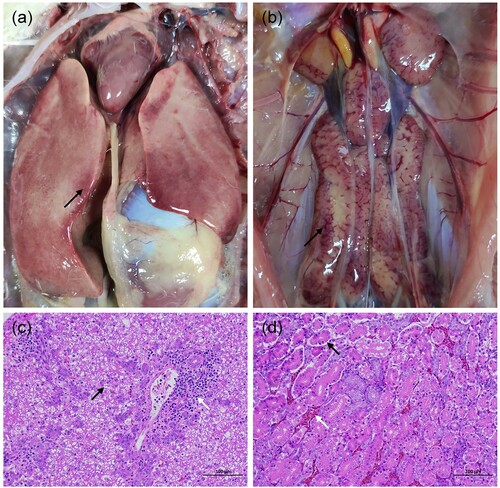
Phylogenetic analysis of hexon gene
In total, 54 clinical samples were analyzed by hexon gene PCR and sequencing. According to the phylogenetic analysis based on the hexon gene with available sequences of 38 avian adenoviruses from GenBank (), 54 clinical samples were classified as DAdV-3, showing high amino acid identities with the reference Chinese DAdV-3 strains (98.7%–100%) and lower similarities with the other reference strains (less than 86.0%).
Virus isolation and identification
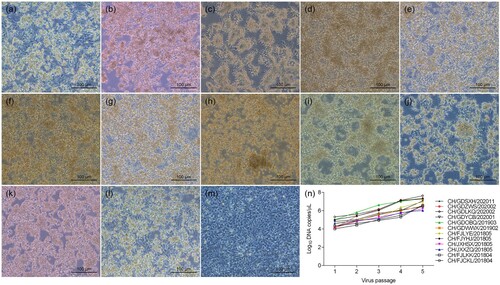
Twelve out of the 23 (52.2%) DAdV-3 strains were successfully isolated from tissue samples collected from diseased Muscovy duck flocks. During serial passage in LMH cell culture, the visible CPE of the 12 DAdV-3 isolates (including CH/FJCKL/201804, CH/FJLKK/201804, CH/JXXZQ/201805, CH/JXHSX/201805, CH/FJYHJ/201805, CH/FJLYE/201805, CH/GDWWX/201902, CH/GDOBQ/201903, CH/GDYCB/202001, CH/GDLKQ/202002, CH/GDZWS/202002, and CH/GDSXH/202011) in LMH cells showed round cells clustered like grapes at 72 h post-infection ((a–l)), as compared with the control LMH cells ((m)), which is similar to the CPE of FAdV-infected cells (Zhang et al., Citation2018; Chen et al., Citation2020). DAdV-3 specific real-time PCR indicated that the viral load continued to increase in LMH cells following five passages ((n)). After the fifth passage, the viral titre reached between 105.5 TCID50/ml and 107.33 TCID50/ml, indicating that the 12 DAdV-3 strains were highly replicative in LMH cells.
Figure 4. Isolation and characterization of DAdV-3 isolates in LMH cell culture. Cytopathic effect of the 12 DAdV-3 strains was observed at 72 h post infection. (a) CH/FJCKL/201804, (b) CH/FJLKK/201804, (c) CH/JXXZQ/201805, (d) CH/JXHSX/201805, (e) CH/FJYHJ/201805, (f) CH/FJLYE/201805, (g) CH/GDWWX/201902, (h) CH/GDOBQ/201903, (i) CH/GDYCB/202001, (j) CH/GDLKQ/202002, (k) CH/GDZWS/202002, (l) CH/GDSXH/202011, (m) Uninfected LMH cells, (n) Viral load of the 12 DAdV-3 strains in LMH cells of different passages.
Phylogenetic analysis of the complete genome of DAdV-3 strains
The whole-genome sequence of 12 DAdV-3 strains was obtained by assembling 17 overlapping sequences. The complete genomes of the 12 DAdV-3 strains were 43,842 nucleotide (nt) in length, and were deposited in GenBank with accession numbers (OK413887, OM117797, OM117798, OM117799, OM117800, OM117801, OM117802, OM117803, OM117804, OM117805, OM117806, and OM117807). We compared the 12 DAdV-3 isolates with 38 other avian adenovirus strains available in GenBank. As shown in , phylogenetic analysis based on the whole genome showed that the 12 DAdV-3 strains clustered within DAdV-3 with 99.8%–100% identity to the reference DAdV-3 strains. Sequence alignments showed that the 12 DAdV-3 isolates shared high amino acid identity in the hexon (99.7%–100%), fibre-1 (97.2%–100%), and fibre-2 (99.8%–100%) with the reference DAdV-3 strains. However, compared to the whole genomes of the reference DAdV-3 strains, the ORF67 gene of nine new isolates (including CH/FJCKL/201804, CH/FJLKK/201804, CH/JXXZQ/201805, CH/JXHSX/201805, CH/FJYHJ/201805, CH/FJLYE/201805, CH/GDWWX/201902, CH/GDYCB/202001, and CH/GDSXH/202011) showed 3ʹ terminal truncation through conversion of codon 45 (GGA) to a stop codon (TGA) ( and ).
Figure 5. Phylogenetic tree constructed based on the whole-genome nucleotide sequences of 12 DAdV-3 strains together with other avian adenoviruses. The 12 DAdV-3 isolates are indicated with black circles.
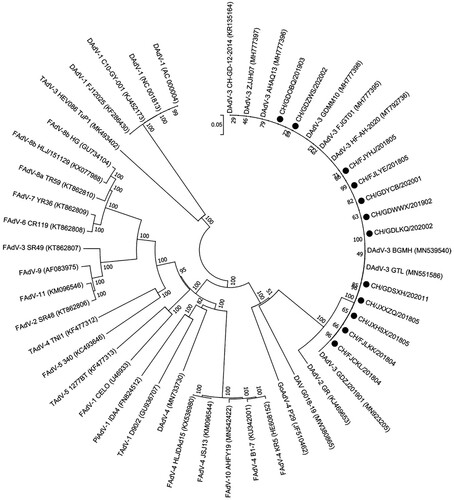
Table 1. Alignment of DAdV-3 amino acids between the 12 new isolates and other representative DAdV-3 strains.
Discussion
In 2014, a novel DAdV-3 strain was firstly isolated from diseased Muscovy ducks in China (Zhang et al., Citation2016). An increasing number of clinical cases of DAdV-3 infection have been reported in China, leading to substantial economic losses in the duck industry (Shi et al., Citation2019; Shi et al., Citation2022). However, little information is available about the epidemiology of DAdV-3 infection. In the present study, the epidemiology of 54 DAdV-3 positive cases collected between 2018 and 2020 in China was investigated. Twelve out of the 23 (52.2%) DAdV-3 strains were isolated on LMH cells. These DAdV-3 strains can be further used for virological and serological assay development, as well as vaccine development.
Previous studies have reported that co-infections with FAdVs and other pathogens are detected frequently in poultry farms (Ojkic et al., Citation2008; Choi et al., Citation2012; Niu et al., Citation2018). The present epidemiological investigation revealed that co-infections of DAdV-3 with other viral or bacterial pathogens were also detected regularly. Of these pathogens, R. anatipestifer was most commonly detected. R. anatipestifer is widely present in ducks, turkeys, geese, and other avian species (Segers et al., Citation1993; Hinz et al., Citation1998), and R. anatipestifer infections cause fibrinous pericarditis, perihepatitis and airsacculitis in all birds (Hess et al., Citation2013; Chikuba et al., Citation2016), which may exacerbate the clinical signs of DAdV-3 in ducks. Moreover, a phylogenetic tree and sequence alignment based on the hexon amino acid revealed that all 54 clinical samples belonged to DAdV-3, and shared 98.7%–100% similarities with DAdV-3 reference strains. However, all clinical samples shared only 86.0%–87.0% hexon amino acid identity with the DAdV-2 GR strain, which is similar to other DAdV-3 strains (Shi et al., Citation2019).
Currently, several systems are reported for DAdV-3 isolation. For example, Zhang et al. (Citation2016), and Shi et al. (Citation2019), reported that primary duck embryo fibroblasts and Muscovy duck embryos are susceptible for isolation of DAdV-3. In addition, a stable Leghorn male hepatoma cell line (LMH cell) is commonly used to isolate some avian adenoviruses, such as FAdV-4 and FAdV-8 (Alexander et al. Citation1998; Yu et al., Citation2018; Chen et al., Citation2020). In this study, we attempted to isolate DAdV-3 from 23 real-time PCR-positive samples using LMH cells. From the 23 samples, 12 DAdV-3 strains were successfully isolated and identified by specific real-time PCR. Clinical sample quality is important for DAdV-3 isolation and the negative samples might have contained very low amounts of infectious virus, making successful isolation difficult. Moreover, Günes et al. (Citation2012) demonstrated that the detection rate of FAdV in samples could be improved considerably by real-time PCR in contrast to virus isolation in cell culture, indicating that real-time PCR is more sensitive than virus isolation.
Phylogenetic analysis of the whole genome demonstrated that the 12 DAdV-3 strains had 99.8%–100% nucleotide identity with the DAdV-3 reference strains in GenBank, and shared 92.3% nucleotide identity with the DAdV-2 GR strain. The most important distinguishing feature between the 12 new isolates and the DAdV-2 GR was that the 12 new isolates had two fibre genes (fibre-1 and fibre-2), whereas the DAdV-2 GR only contained one fibre gene (Marek et al., Citation2014), which is similar to other Chinese DAdV-3 isolates (Zhang et al., Citation2016; Shi et al., Citation2019). Moreover, sequence alignment of the whole genome showed that nine out of the 12 (75%) DAdV-3 isolates had truncated ORF67, which is similar to observations in some DAdV-3 strains (Shi et al., Citation2022), indicating that these mutant strains have become the dominant strains in China.
In summary, DAdV-3 infection was prevalent in Muscovy duck flocks in three provinces in China during 2018-2020, and mutant strains (truncated ORF67) are currently the predominant strains causing swelling and haemorrhagic liver and kidney in Muscovy ducks. To the best of our knowledge, this is the first study providing epidemiological characteristics of DAdV-3 in ducks in China.
Ethics statement
All bird experiments were carried out strictly in accordance with the “Guidelines for Experimental birds” of the Ministry of Science and Technology (Beijing, China). Permission was issued for the field studies, and all of the owners of the farms gave informed consent to perform the study on this site. The bird experiment protocol was approved by the Committee of the Ethics of bird Experiments at the South China Agricultural University (approval ID: SYXK-2014-0136).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alexander, H.S., Huber, P., Cao, J., Krell, P.J. & Nagy, E. (1998). Growth characteristics of fowl adenovirus type 8 in a chicken hepatoma cell line. Journal of Virological Methods, 74, 9–14.
- Cha, S.Y., Kang, M., Park, C.K., Choi, K.S. & Jang, H.K. (2013). Epidemiology of egg drop syndrome virus in ducks from South Korea. Poultry Science, 92, 1783–1789.
- Chen, H., Dou, Y., Zheng, X., Tang, Y., Zhang, M., Zhang, Y., Wang, Z. & Diao, Y. (2017). Hydropericardium hepatitis syndrome emerged in Cherry Valley ducks in China. Transboundary and Emerging Diseases, 64, 1262–1267.
- Chen, L., Yin, L., Peng, P., Zhou, Q., Du, Y., Zhang, Y., Xue, C. & Cao, Y. (2020). Isolation and characterization of a novel fowl adenovirus serotype 8a strain from China. Virologica Sinica, 35, 517–527.
- Chikuba, T., Uehara, H., Fumikura, S., Takahashi, K., Suzuki, Y., Hoshinoo, K. & Yamamoto, Y. (2016). Riemerella anatipestifer infection in domestic ducks in Japan, 2014. Journal of Veterinary Medical Science, 78, 1635–1638.
- Choi, K.S., Kye, S.J., Kim, J.Y., Jeon, W.J., Lee, E.K., Park, K.Y. & Sung, H.W. (2012). Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poultry Science, 91, 2502–2506.
- Günes, A., Marek, A., Grafl, B., Berger, E. & Hess, M. (2012). Real-time PCR assay for universal detection and quantitation of all five species of fowl adenoviruses (FAdV-A to FAdV-E). Journal of Virological Methods, 183, 147–153.
- Hansen, W.R., Brown, S.E., Nashold, S.W. & Knudson, D.L. (1999). Identification of duck plague virus by polymerase chain reaction. Avian Diseases, 43, 106–115.
- Harrach, B. (2014). Adenoviruses: general features. In M. Caplan, R. Mitchell, R. Bradshaw & L. McManus (Eds.), Reference module in biomedical sciences (pp. 1–9). Amsterdam: Elsevier.
- Harrach, B., Benkő, M., Both, G.W., Brown, M., Davison, A.J., Echavarra, M., Hess, M., Jones, M.S., Kajon, A., Lehmkuhl, H.D., Mautner, V., Mittal, S.K. & Wadell, G. (2011). Family adenoviridae. In A.M.Q. King, M.J. Adams & E.J. Lefkowitz (Eds.), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses (pp. 125–141). San Diego: Elsevier.
- Hess, C., Enichlmayr, H., Jandreski-Cvetkovic, D., Liebhart, D., Bilic, I. & Hess, M. (2013). Riemerella anatipestifer outbreaks in commercial goose flocks and identification of isolates by MALDI-TOF mass spectrometry. Avian Pathology, 42, 151–156.
- Hinz, K.H., Ryll, M., Kohler, B. & Glunder, G. (1998). Phenotypic characteristics of Riemerella anatipestifer and similar micro-organisms from various hosts. Avian Pathology, 27, 33–42.
- Hu, Q., Tu, J., Han, X., Zhu, Y., Ding, C. & Yu, S. (2011). Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli, and Salmonella enterica simultaneously from ducks. Journal of Microbiological Methods, 87, 64–69.
- Huang, Y., Kang, H., Dong, J., Li, L., Zhang, J., Sun, J., Zhang, J. & Sun, M. (2020). Isolation and partial genetic characterization of a new duck adenovirus in China. Veterinary Microbiology, 247, 108775.
- Kumar, S., Stecher, G. & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
- Lin, S., Wang, S., Cheng, X., Xiao, S., Chen, X., Chen, S., Chen, S. & Yu, F. (2019). Development of a duplex SYBR Green I-based quantitative real-time PCR assay for the rapid differentiation of goose and Muscovy duck parvoviruses. Virology Journal, 16, 6.
- Liu, R., Chen, C., Huang, Y., Cheng, L., Lu, R., Fu, G., Shi, S., Chen, H., Wan, C., Fu, Q. & Lin, J. (2018). Microbiological identification and analysis of waterfowl livers collected from backyard farms in southern China. Journal of Veterinary Medical Science, 80, 667–671.
- Marek, A., Kajan, G.L., Kosiol, C., Harrach, B., Schlotterer, C. & Hess, M. (2014). Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology, 462-463, 107–114.
- Niu, Y., Sun, Q., Zhang, G., Sun, W., Liu, X., Xiao, Y., Shang, Y. & Liu, S. (2018). Epidemiological investigation of outbreaks of fowl adenovirus infections in commercial chickens in China. Transboundary and Emerging Diseases, 65, e121–e126.
- Ojkic, D., Martin, E., Swinton, J., Vaillancourt, J.P., Boulianne, M. & Gomis, S. (2008). Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathology, 37, 95–100.
- Pallister, J., Wright, P.J. & Sheppard, M. (1996). A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. Journal of Virology, 70, 5115–5122.
- Pan, Q., Liu, L., Wang, Y., Zhang, Y., Qi, X., Liu, C., Gao, Y., Wang, X. & Cui, H. (2017). The first whole genome sequence and pathogenicity characterization of a fowl adenovirus 4 isolated from ducks associated with inclusion body hepatitis and hydropericardium syndrome. Avian Pathology, 46, 571–578.
- Reed, L.J. & Muench, H. (1938). A simple method of estimating fifty percent endpoints. American Journal of Hygiene, 27, 493–497.
- Russell, W.C. (2009). Adenoviruses: update on structure and function. Journal of General Virology, 90, 1–20.
- Segers, P., Mannheim, W., Vancanneyt, M., De Brandt, K., Hinz, K.H., Kersters, K. & Vandamme, P. (1993). Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. International Journal of Systematic Bacteriology, 43, 768–776.
- Shi, S., Liu, R., Wan, C., Cheng, L., Chen, Z., Fu, G., Chen, H., Fu, Q. & Huang, Y. (2019). Isolation and characterization of duck adenovirus 3 circulating in China. Archives of Virology, 164, 847–851.
- Shi, X., Zhang, X., Sun, H., Wei, C., Liu, Y., Luo, J., Wang, X., Chen, Z. & Chen, H. (2022). Isolation and pathogenic characterization of duck adenovirus 3 mutant circulating in China. Poultry Science, 101, 101564.
- Su, X.N., Liu, J.J., Zhou, Q.F., Zhang, X.H., Zhao, L.C., Xie, Q.M., Chen, W.G. & Chen, F. (2017). Isolation and genetic characterization of a novel adeno-associated virus from Muscovy ducks in China. Poultry Science, 96, 3867–3871.
- Wan, C., Chen, C., Cheng, L., Fu, G., Shi, S., Liu, R., Chen, H., Fu, Q. & Huang, Y. (2018). Development of a TaqMan-based real-time PCR for detecting duck adenovirus 3. Journal of Virological Methods, 261, 86–90.
- Wang, Y., Li, Y., Yang, C., Hui, L., Han, Q., Ma, L., Wang, Q., Yang, G. & Liu, Z. (2013). Development and application of a universal Taqman real-time PCR for quantitation of duck hepatitis B virus DNA. Journal of Virological Methods, 191, 41–47.
- Yu, X., Wang, Z., Chen, H., Niu, X., Dou, Y., Yang, J., Tang, Y. & Diao, Y. (2018). Serological and pathogenic analyses of fowl adenovirus serotype 4 (FAdV-4) strain in Muscovy ducks. Frontiers in Microbiology, 9, 1163.
- Zhang, J., Zou, Z., Huang, K., Lin, X., Chen, H. & Jin, M. (2018). Insights into leghorn male hepatocellular cells response to fowl adenovirus serotype 4 infection by transcriptome analysis. Veterinary Microbiology, 214, 65–74.
- Zhang, R., Xia, L., Chen, J., Gong, Y., Zhang, L., Li, P., Liu, H., Xie, Z. & Jiang, S. (2017). Molecular epidemiology and genetic diversity of duck hepatitis A virus type 3 in Shandong province of China, 2012–2014. Acta Virologica, 61, 463–472.
- Zhang, X., Zhong, Y., Zhou, Z., Liu, Y., Zhang, H., Chen, F., Chen, W. & Xie, Q. (2016). Molecular characterization, phylogeny analysis and pathogenicity of a Muscovy duck adenovirus strain isolated in China in 2014. Virology, 493, 12–21.
- Zheng, M., Chen, X., Wang, S., Wang, J., Huang, M., Xiao, S., Cheng, X., Chen, S., Chen, X., Lin, F. & Chen, S. (2020). A TaqMan-MGB real-time RT-PCR assay with an internal amplification control for rapid detection of Muscovy duck reovirus. Molecular and Cellular Probes, 52, 101575.