 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
We study Markov evolution algebras, that is, evolution algebras having Markov structure matrices. We first consider the discrete-time case, and delve into their algebraic structure for later application to continuous-time Markov evolution algebras that arise defined by standard stochastic semigroups. The study of embeddable Markov evolution algebras, that is, of Markov evolution algebras with structure matrices existing within standard stochastic semigroups is then equivalent to the embedding problem for Markov matrices.
1. Introduction
An evolution algebra is an n-dimensional (real) algebra with basis
and multiplication:
Such a basis
is called natural basis and the matrix
the structure matrix of
[Citation1,Citation2] ( evolution matrix in [Citation3]).
Evolution algebras were introduced in [Citation1] (see also [Citation2]) and have been exhaustively studied over the last years. See, for instance, [Citation3–6] and references therein for a review of some of the main results achieved on this topic. These algebras are related to a wide variety of mathematical subjects, including Markov chains and dynamical systems. The relationship between evolution algebras and homogeneous discrete-time Markov chains was settled in [Citation1, Theorem 16, p. 54]. Markov evolution algebras are real evolution algebras whose structure matrix A is nonnegative and has row sums equal to 1 (i.e. A is a Markov matrix).
In his seminal work [Citation1], Tian posed as one of the most fruitful further topics of research the development of the theory of continuous evolution algebras and their connection to continuous-time Markov processes. Tian outlined continuous evolution algebras to be evolution algebras with multiplication, w.r.t. a natural basis :
for differentiable functions aij(t), i, j = 1, …, n [Citation1, Section 6.2.4, p. 115].
Chains of evolution algebras (CEAs), introduced in [Citation7], can be understood as the first approach to continuous-time evolution algebras, as they consist of families of evolution algebras whose structure matrices satisfy Chapman–Kolmogorov-type equations [Citation7, Theorem 2.5]. Continuity or differentiability is, however, not required.
With the further aim of settling a theory of continuous evolution algebras, in this paper, we put the focus on Markov evolution algebras. First, we review the discrete-time case already considered in [Citation1]. Here, taking advantage of recently obtained results on general evolution algebras, we improve our knowledge on these structures, for later application to the continuous-time case. In the second part of the paper, we introduce the notion of continuous-time Markov evolution algebra as the first step to a more general theory of continuous evolution algebras. Our study of continuous-time Markov evolution algebras connects these algebras to the embedding problem for Markov matrices [Citation8].
This paper is organized as follows. After this introductory section, in Section 2, we revisit Tian's notion of Markov evolution algebra, characterized as those evolution algebras whose structure matrix is precisely a (real) Markov matrix (i.e. nonnegative and row stochastic), therefore defining a homogeneous discrete-time Markov chain. In this section, we address the elementary properties of this class of evolution algebras, such as the (non)existence of nilpotent or idempotent elements. We also study the behaviour of their evolution operator, and show that results in [Citation9] can be seen as a generalization for the ergodic theorems for Markov evolution algebras having primitive structure matrices [Citation10].
In the third section, we take advantage of the notion of basic ideal [Citation11] to address a decomposition of Markov evolution algebras into simple basic ideals together to an additional component gathering the inessential behaviour within the Markov evolution algebra. This decomposition, similar to the semi-direct sum decomposition proposed in [Citation1, 4.4, p. 76], stems quite straightforwardly when considering the notion of an essential class of states (and also that of incidence matrix) for nonnegative matrices. It also comes out the suitability of the notion of basic ideal to study Markov evolution algebras. Section 4 is devoted to examples. The different Markov evolution algebras considered in this section, stemming them all mainly from Biology, allow us to illustrate different algebraic properties of Markov evolution algebras.
In Section 5, we recall some basic facts on continuous-time Markov chains [Citation12,Citation13], prior to introducing in Section 6 continuous-time Markov evolution algebras to be families of (Markov) evolution algebras whose structure matrices {A(t)}t≥0 form standard stochastic semigroups, and therefore are solutions of Kolmogorov differential equations. Being, for all t ≥ 0, the structure matrix of
of the form A(t) = etQ, it comes out that all such evolution algebras should be not only nondegenerate but also perfect, posing therefore the question of identifying those evolution algebras that can be realized this way. This leads us to consider the embedding problem.
The embedding problem copes with the problem of given a Markov matrix P to determine the existence of a rate matrix Q such that P = eQ and etQ is Markov for all t ≥ 0. The existence and uniqueness (i.e. the identifiability) of the Markov generator Q is related to the spectral properties of P and to the noninjectivity of the principal logarithm of matrices [Citation8]. See [Citation14,Citation15] and references therein for recent results on the embedding problem in phylogenetics.
The study of CT-Markov evolution algebras leads naturally to consider when a Markov evolution algebra (its structure matrix) can exist within a standard stochastic semigroup. To do this, we consider the embeddability of discrete-time Markov evolution algebras, and although determining families of embeddable matrices it not a goal pursued here, some such characterizations will be applied in Section 7 to obtain Markov evolution matrices realizable within CT Markov processes.
2. Markov evolution algebras
Let be the field of real numbers. In what follows, all algebraic objects will be defined over finite-dimensional real vector spaces. Matrices and vectors will be denoted boldface.
Evolution algebras are a class of commutative algebras, which are not necessarily associative nor power-associative [Citation1, Corollary 1, p. 20]. Only nonzero trivial evolution algebras with structure matrix A = diag(λ1, …, λn), (λi ≠ 0, for all i) are unital [Citation1, Proposition 1, p. 22].
Definition 2.1
[[Citation1, 4.1.2, p. 54]] Let be a finite-dimensional real algebra (
). We say that
is a Markov evolution algebra if it admits a basis
and multiplication:
(i.e.
is an evolution algebra), such that its structure matrix
is a nonnegative row stochastic matrix, i.e. a (real) Markov matrix.
The structure matrix A of any Markov evolution algebra can be seen as the transition probability matrix of a homogeneous discrete-time (HDT) Markov chain on a finite set of states Λ = {1, …, n}, under the obvious identification . Within the genetic non-Mendelian framework, where evolution algebras first arose, one can write
where aij = P(ej| ei) denotes the probability for a type j individual to arise from self-replication of a type i individual.
We refer the reader to [Citation10,Citation13] for basic results on HDT Markov processes.
Definition 2.2
A discrete-time stochastic process , where Xk is a discrete random variable defined on a (finite) state space Λ = {1, …, n}, is a Markov chain if the Markov property
where ik ∈ {1, …, n}, holds for all k = 0, 1, …. A discrete-time Markov chain is homogeneous (HDT) if the transition probabilities do not depend on (time) k, P(Xk = ik| Xk−1 = ik−1) = P(Xk+l = ik| Xk+l−1 = ik−1), for all l = 0, 1, …. We denote the transition probabilities of a HDT Markov chain by aij = P(Xk = j| Xk−1 = i), k = 0, 1, ….
The relation between Markov evolution algebras and HDT Markov chains was settled in [Citation1, Theorem 16, p. 54].
Theorem 2.3
[Citation1, Theorem 16, p. 54]
For each homogeneous Markov chain X there is an evolution algebra MX whose structural constants are transition probabilities, and whose generator set is the state space of the Markov chain.
Examples 2.4
Let be an evolution algebra with an natural basis
.
The only trivial Markov evolution algebra has A = In, that is,
, for all i = 1, …, n. In this particular case,
is unital with unit element
.
An evolution algebra of permutations
is Markov if and only if ai = 1, i = 1, …, n. Then A equals the permutation matrix of π ∈ Sn [Citation16].
Let
be the evolution algebra corresponding to permutations π, τ (π ≠ τ ∈ Sn), with multiplication [Citation17]:
Then
is Markov if and only if aiπ(i), aiτ(i) ≥ 0 and aiπ(i) + aiτ(i) = 1 for all i = 1, …, n.
Evolution algebras
and
[Citation7] are Markov evolution algebras:
Here,
and
. Algebras
,
and
in the classification of 2-dimensional real evolution algebras [Citation18, Theorem 1] are also Markov evolution algebras, with structure matrices:
Notice now
, whereas
. We recall here that the rank
of an evolution algebra
equals
.
Remark 2.1
As many other properties of evolution algebras, being Markov may depend on the natural basis. Therefore, natural basis (as evolution algebras) of Markov evolution algebras need to be assumed to be Markov-natural (M-natural, for short), that is, to be bases giving rise to Markov structure matrices. To illustrate this, let us consider the 2-dimensional evolution algebra (see Example 2.4(iv)) with natural basis
and multiplication
. The structure matrix of
is then:
Consider now a second (natural) basis
, where f1 = e1 + e2 and f2 = e1 − e2. Then
and
giving rise to the structure matrix
which is clearly not row stochastic. Hence
is M-natural, but
is not.
Example 2.5
Consider the evolution algebras induced by the cycle graph with five vertices C5 and by the random walk on C5 [Citation19, Example 2.5]:
Both
and
are evolution algebras w.r.t.
, but only
is Markov. However,
by [Citation20, Theorem 3.2(i)]. The case of the wheel graph Wn (n>4) is similar, with
being Markov, but
not Markov. However,
as evolution algebras [Citation20, Proposition 3.5].
In what follows, natural bases of Markov evolution algebras will be assumed to be M-natural basis and to remain fixed throughout.
M-natural bases are normalized natural bases ( for all i = 1, …, n w.r.t. the l1-norm) and unital (
). Thus Markov evolution algebras are normed algebras w.r.t.
and therefore, by finite-dimensionality, Banach evolution algebras [Citation9, Corollary 2.5] and [Citation1, Chapter I.3.3, p. 36].
The notion of graph attached to an evolution algebra
relative to a natural basis
was introduced in [Citation6, Definition 2.2]. Recall (i, j) ∈ E if and only if aij ≠ 0. This corresponds to the usual notion of (weighted) digraph G(A) associated to its structure matrix A and therefore implies having state i leading to state j (i → j) in the Markov chain, since i → j if and only if
for some integer k ≥ 1 [Citation10, p. 12].
Remark 2.2
Baric evolution algebras were characterized in [Citation7, Theorem 3.2] as follows: An n-dimensional real evolution algebra is baric if and only if there is a column
of its structure matrix A with
and
for all i ≠ i0. The weight map is then defined by
, for all
. Moreover, see [Citation7, Corollary 3.3],
has exactly as many weight functions as columns
of the structure matrix A satisfy the given condition, j = i1, …, im, m ≤ n.
Proposition 2.6
Let be a Markov evolution algebra. If
is a baric algebra, then the underlying HDT Markov chain is reducible.
Proof.
If is baric, by [Citation7, Theorem 3.2], its structure matrix A contains a column
such that
, which amounts to
for any i ≠ i0, meaning that i0 cannot be reached from any other state i, and therefore implying that the underlying HDT Markov chain is reducible. This follows from the fact all entries in the i0th column of
vanishes, but
.
Remark 2.3
We note that reducibility is, however, not enough for ensuring the existence of characters. Indeed, consider the Markov evolution algebra with structure matrix
and reducible underlying Markov chain, having two (obviously disjoint) essential classes. Clearly, [Citation7, Theorem 3.2] is not satisfied, and therefore
is not baric.
Graphically (also equivalently, in terms of the Markov chain states topology), a Markov evolution algebra is baric if and only if there exists a state i0 ∈ {1, …, n} such that i0 → i0 ( has a loop at i0) and
, for any i ≠ i0, that is i0 is a source. The number of different weight maps is then given by the number of sources of
, with a loop at them. The existence of sources, being a requirement for that of characters, leads us to the following result.
Corollary 2.7
Let be a Markov evolution algebra. If
is strongly connected, then
has no nonzero character (
is not a baric algebra).
Proposition 2.8
Markov evolution algebras are not nil (nor nilpotent).
Proof.
Let be a Markov evolution algebra with M-natural basis
. Then no reordering of the elements of
can produce an strictly upper triangular matrix (as all row totals remain equal to 1). The result now follows from [Citation5, Theorem 2.7].
Proposition 2.9
Markov evolution algebras have no nonzero absolute nilpotent elements (i.e. elements such that x2 = 0).
Proof.
Let and write
. Assume now x is a nonzero absolute nilpotent element and take i0 ∈ {1, …, n} such that
. Since A is nonnegative, it then follows that
for all j = 1, …, n, contradicting the row stochasticity of A.
Proposition 2.10
Let be a Markov evolution algebra. Then the evolution element
is idempotent if and only if A is a doubly stochastic matrix.
Proof.
We note that . Thus, since A is nonnegative, e2 = e amounts to have
for all j = 1, …, n, that is, to A also have column sums equal to 1.
Spectral theory of nonnegative matrices, including Markov matrices, has been broadly studied [Citation10,Citation13]. A nonnegative n × n matrix A (A ≥ 0 i.e. aij ≥ 0 for all i, j = 1, …, n) is positive (A > 0) if aij > 0 for all i, j = 1, …, n; and primitive if Ak > 0 for some positive integer k. Irreducibility means that for every pair i, j ∈ {1, …, n} there exists a positive integer k such that , the (i, j)th entry of Ak, is (strictly) positive. Eigenvalues of primitive and irreducible Markov matrices follow from Perron–Frobenius Theorem [Citation10, Theorem 1.1, Theorem 1.5]. If A is primitive and Markov, then A has eigenvalue λ = 1 with algebraic and geometric multiplicity 1, and |λi| < 1 for any other eigenvalue λi ≠ 1 of A. If A is irreducible, but not primitive, then |λi| ≤ 1, and, in general, algebraic and geometric multiplicities of λ = 1 are given by the number of communication classes of A [Citation13, pp. 197–198]. Equivalent results can be found in [Citation1, 4.3].
The analytic study of evolution algebras, carried on in [Citation9] and focused on the ergodic behaviour of evolution operators of finite-dimensional evolution algebras, can be understood as a generalization of previously mentioned results in the case of primitive nonnegative matrices.
Definition 2.11
The evolution operator of an evolution algebra (w.r.t.
) is defined as the (left) multiplication L(x) = ex by the evolution element
.
The evolution operator L of a Markov evolution algebra leaves invariant the (n − 1)-dimensional simplex Δ(n−1) that can be understood as the set of probability distributions of the underlying (genetic, dynamical,…) system.
Remark 2.4
Let a be an element of a finite-dimensional commutative real algebra , and let La(x) = ax, for all
, be the left multiplication by the element a. The m-spectrum of a is
where as usual,
denotes the spectrum of La (as linear map). The m-spectral radius of a is
Given λ ∈ σm(a), the multiplicative a-index of λ, denoted ν(λ, a) = ν(λ, La), is the size of the largest Jordan block corresponding to λ in the canonical Jordan normal block matrix associated to La (with ν(λ, a) = 0 if
). We refer the reader to [Citation9, Definition 4.3, Definition 4.4, Theorem 4.13, Definition 4.14] for more accurate details on the m-spectrum, m-spectral radius and multiplicative index of the elements of evolution algebras. We only note here that, despite the differences with the notation used in [Citation9], the eigenvalues of the evolution operator L = Le are exactly those of the structure matrix A of
[Citation9, Equation 4.1, p. 120].
Theorem 2.12
Let e be the evolution element of a Markov evolution algebra (w.r.t. a M-natural basis
) with primitive structure matrix A. Then:
and ρ(e) = 1.
The multiplicative e-index of λ = 1 is ν(1, e) = 1.
e is an equilibrium generator of
, equivalently
reaches
-equilibrium, that is
converges for all
. (See equivalent conditions in [Citation9, Proposition 4.7].)
Proof.
(i) Follows from [Citation9, Proposition 4.5] taking into account Perron–Frobenius Theorem for primitive Markov matrices. For (ii), it suffices to recall that the algebraic and geometric multiplicity of λ = 1 for A is one (see more details in [Citation9, Proposition 4.13]). Finally, (iii) is [Citation9, Corollary 4.17].
Remark 2.5
In this setting, we have dim(ker(A − In)) = ma(1, A) = mg(1, A) = 1 = rank(lim k→∞Ak) [Citation9, Proposition 4.10, Proposition 4.13]. Note the last equality also follows from the Ergodic Theorem for Primitive Markov Chains [Citation10, Theorem 4.2], since then Ak → 1vT, where v denotes the unique stationary distribution of the Markov chain, and convergence is elementwise. Therefore results in [Citation9, Proposition 4.7] on the ergodicity of multiplication operators of the form can be understood as a generalization of the Ergodic Theorem for Primitive Markov Chains for arbitrary finite-dimensional evolution algebras.
Example 2.13
Consider a 2-dimensional Markov evolution algebra with positive (hence primitive) structure matrix
for some a, b ∈ (0, 1). Then σ(A) = {1, 1 − a − b} and dim(ker(A − In)) = ma(1, A) = mg(1, A) = 1. A classical result states that 1 = (1, 1)T is a right eigenvector of A of eigenvalue 1 with stationary probability distribution
, and
has rank one. Thus, given
, we have
, with x = (x1, x2)T.
3. The structure of Markov evolution algebras
In this section, we delve into the structure of Markov evolution algebras. To do this basic ideals are considered. A proper ideal I of an evolution algebra is an i-basic ideal w.r.t. a natural basis
if I is generated by i vectors of
[Citation11, Definition 2.1]. We recall here, that for any finite-dimensional perfect evolution algebra (
), simplicity and basic simplicity are equivalent [Citation11, Proposition 2.7].
Definition 3.1
[Citation3, Definition 2.16]
An evolution algebra is nondegenerate if it has a natural basis
such that
for all i = 1, …, n. Nondegeneracy is equivalent to
[Citation3, Corollary 2.19].
Proposition 3.2
Markov evolution algebras are nondegenerate.
Proof.
Recall first [Citation6, Lemma 2.7]. Hence to prove that a Markov evolution algebra is nondegenerate, it suffices to check
, for all i = 1, …, n. But this follows from Proposition 2.9 on the existence of no nonzero absolute nilpotent elements in Markov evolution algebras. Indeed, note that
would imply that the ith row Ai: = (ai1, …, ain)T of the structure matrix A of
vanishes, contradicting the row stochasticity of A.
Proposition 3.3
Let be a Markov evolution algebra. Then
is indecomposable (there exist no nonzero ideals I and J such that
) if and only if
is strongly connected.
Proof.
It follows from [Citation6, Proposition 2.8], as a result of Proposition 3.2.
Ideals considered in Proposition 3.3 have their usual meaning, despite the different notions of (evolution) ideals found in the literature (see , for instance, [Citation1,Citation3]). A similar result follows from [Citation3, Lemma 5.2, Corollary 5.8], as a consequence of nondegenerancy of Markov evolution algebras (Proposition 3.2).
Remark 3.1
A Markov evolution algebra is perfect if and only if its structure matrix A is nonsingular (det(A) ≠ 0) [Citation6, Proposition 4.2]. This follows into natural bases to be unique up to permutation and scalar multiplication [Citation6, Corollary 4.7]. A similar result holds for M-natural basis, since if
is M-natural, so is
for all σ ∈ Sn.
Following the previous section, in what follows, we consider to be a Markov evolution algebra with structure matrix A w.r.t. a M-natural basis
, and identify
in the obvious way, where Λ = {1, …, n} is the state set of the underlying HDT Markov chain.
Remark 3.2
We refer to [Citation10, p. 12] for the notion of essential and inessential classes of states of nonnegative matrices, and to [Citation12, II.2.3] for the notion of communication class of states of Markov chains. We note that any essential class forms a communication class, but the converse is not true, as communication classes may not be closed [Citation12, Definition 2.9]. Indeed, states in an essential class communicate to each other, but cannot lead to any other state outside their class. However, the equivalence relation defining communication classes [Citation12, Definition 2.7] does not exclude the existence of states leading to states outside it.
Lemma 3.4
Let be a Markov evolution algebra with M-natural basis
. Then, under
any closed subset of states of Λ generates a basic ideal of
and, conversely, any basic ideal defines a closed subset of Λ. This ideal is basic simple if and only if the closed subset forms an essential class.
Proof.
Assume, after relabelling if necessary, C = {1, …, r} ⊂ Λ is closed. Then for all 1 ≤ i ≤ r < j ≤ n, and any positive integer
. This implies
, for every i ∈ C. Hence I is a basic ideal of
. Conversely, let I be a basic ideal of
, and write
, with C ⊂ Λ. Then, for all i ∈ C,
implies aij = 0 for all
, and also
for all
. Hence C is closed.
The last statement is clear.
Theorem 3.5
Let be a Markov evolution algebra with structure matrix A w.r.t. a M-natural basis
. Then, suitably relabelling the elements of
:
A has a block structure:
where Ak is a nk × nk matrix, for k = 1, …, r, and
(disjoint union), with Λk = {n1 + · · · + nk−1 + 1, …, n1 + · · · + nk} for k = 1, …, r + 1, and n = n1 + · · · + nr+1.
is an nk-basic ideal of
with M-natural basis
and structure matrix Ak, for all 1 ≤ k ≤ r. Moreover, IkIs = 0 for all k ≠ s = 1, …, r.
Ideals Ik are basic simple, for all k = 1, …, r.
where
is a proper (n1 + · · · + ns)-basic ideal of
, for all s = 1, …, r, is a filtration of
.
admits a grading
with
and
for 2 ≤ s ≤ r + 1. Moreover,
for 1 ≤ s ≤ r.
Proof.
(i) The block structure of A, after a suitable index relabelling, arises from the classification into essential and inessential classes of the indices of any nonnegative matrix and it corresponds to the so-called canonical form of the nonnegative matrix [Citation10, p. 14]. It is then straightforward to check (ii), i.e. that Ik is an nk-basic ideal with M-natural basis and structure matrix Ak, for all 1 ≤ k ≤ r. The orthogonality of these ideals is clear.
Next, (iii) follows from Lemma 3.4 and, finally, statements (iv)–(v) are standard.
The threefold identification between elements of M-natural bases of Markov evolution algebras, indices of their (nonnegative) structure matrices and states of the underlying HDT Markov chains, is the cornerstone of the above decomposition into minimal basic ideals, together to an additional term gathering the inessential (transient) behaviour within the Markov evolution algebras. We recall here that classification into (in)essential classes only depends on the vanishing or non-vanishing of the (nonnegative) entries aij. Thus any two Markov evolution algebras with the same incidence matrix (the matrix that arises when replacing all strictly positive matrix entries of the structure matrix by ones) have the same decomposition. This fact can also be seen as a result of both having the same (un-weighted) digraph.
4. Examples
Although evolution algebras were firstly introduced in connection to non-Mendelian genetics, some of their most recent examples arise from those areas they turned out to be related to [[Citation20, 2.1], [Citation1, 4.4.7]]. In this section, we exploit some well-known Markov processes modelling different stochastic processes to define the corresponding Markov evolution algebras.
4.1. Birth and death processes
Consider a birth and death process on {0, 1…, N} with absorbing states 0 and N. The (N + 1) × (N + 1) transition matrix is
where we have (p + q = 1, p, q ≥ 0):
and, additionally, a00 = 1 = aNN [Citation12, Example 2.3]. Let
be the (N + 1)-dimensional Markov evolution algebra with structure matrix A w.r.t.
. Clearly det(A) = 0 and, the decomposition of the state space Λ = {0, 1…, N} of the underlying HDT Markov chain into three (disjoint) communication classes
(see Remark 3.2) proves that
is not simple as it contains two 1-dimensional (basic) ideals
and
, both corresponding to absorbing states. Note, however, that
is not a (basic) ideal, since the communication class {1, 2, …, N − 1} is not closed. This fact is easily illustrated by the digraph
, that in case when N = 3 is given by
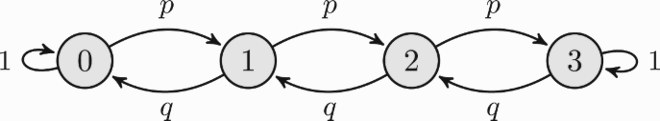
This Markov evolution algebra provides an alternative example of non-perfect finite-dimensional evolution algebra with non-unique maximal basic ideals [Citation11, Example 2.6]. It is also clear that
is not baric (see Remarks 2.2 and 2.3).
General birth and death processes [Citation12, 3.6] do provide more general examples of Markov evolution algebras, with properties such as being perfect, simple or baric depending on birth and death probabilities.
4.2. Greenwood model
Greenwood model is a simplified version of a bivariate Markov chain for binomial epidemic models. Let , being Sk and Ik discrete random variables for, respectively, the number of susceptible and infected individuals at time k. In this model Sk = Sk+1 + Ik+1, that is, susceptible individuals at time k are either still susceptible or already infected at time k + 1, as no births nor deaths are assumed [Citation21]. In Greenwood model
where 1−p is the probability of contact resulting in infection. Assuming I0 > 0, the state space is Λ = {0, 1, …, s0} with S0 = s0 > 0 being the maximal number of possible infected individuals [Citation12, 3.10.1]. The transition matrix is
giving
, for all i = 0, 1, …, s0.
The corresponding n = (s0 + 1)-dimensional Markov evolution algebra is perfect (
), but not simple, since
is an (i + 1)-dimensional (basic) ideal of
, for all i = 0, …, s0 [Citation11, Proposition 2.7]. Assuming s0 = 3 and following the terminology introduced in [?],
provides an example of 4-dimensional perfect non-simple evolution algebra containing a 3-basic ideal which has a 2-basic ideal, not satisfying classification condition (3, 2, 3).
4.3. Jukes–Cantor model
The Jukes–Cantor model for molecular evolution has doubly stochastic transition matrix:
where for i, j = A, G, C, T (DNA bases) aij = P(Xk+1 = j| Xk = i) = a, i ≠ j. Moreover, det(A) = (1 − 4a)3 and σ(A) = {1, 1 − 4a}. Thus A is a positive, hence irreducible, matrix if a ∈ (0, 1/3), and for a ∈ (0, 1/3) − {1/4}, the corresponding evolution algebra
with structure matrix A is a 4-dimensional simple (perfect) Markov algebra, and
is strongly connected. We recall here that Jukes-Cantor model is a particular case of Kimura 3ST model with additional symmetries (see, for instance, [Citation22, Definition 2.1]).
5. Continuous-time Markov chains
Let {X(t)| t ∈ [0, ∞)} be discrete random variables with values in a finite set Λ = {1, …, n}, and continuous index set t ∈ [0, ∞). We refer to [Citation12,Citation13] for background on these processes.
Definition 5.1
A continuous-time Markov chain (CTMC) is a stochastic process {X(t)}t≥0 satisfying the Markov property, i.e. for any sequence 0 ≤ t0 < t1 < … < tk < tk+1, P(X(tk+1) = ik+1| X(t0) = i0, …, X(tk) = ik) = P(X(tk+1) = ik+1| X(tk) = ik).
Assuming transition probabilities pij(t, s) = P(X(t) = j| X(s) = i), for s<t, only depend on the time interval length t−s (homogeneity), we write pij(t) = P(X(t) = j| X(0) = i). Transition probability matrices are nonnegative row stochastic for all t ≥ 0 (i.e. Markov matrices), and satisfy Chapman–Kolmogorov equation P(t + s) = P(t)P (s), for all s, t ≥ 0. Also P(0) = In, since pij(0) = P(X(0) = j| X(0) = i) = δij, i, j = 1, …, n. Functions pij(t) = P(X(t) = j| X(0) = i), for all t ≥ 0, are the transition functions of the process, and the set {P(t)}t≥0 is the transition or stochastic semigroup of the CTMC on Λ = {1, …, n}.
Definition 5.2
[Citation13, Chapter 8, Section 2.2]
A family {P(t)}t≥0 of (n × n) real matrices forms a (finite) stochastic semigroup on a finite Λ = {1, …, n}, if for each t, s ≥ 0:
P(t) is a Markov matrix.
P(0) = In.
P(t + s) = P(t)P(s) (Chapman–Kolmogorov equation or semigroup property).
A stochastic semigroup is called standard if:
, componentwise.
Remark 5.1
As |Λ| = n is finite, continuity at origin (t = 0) of stochastic semigroups ensures continuity at any other time t ≥ 0, as well as differentiability at the origin. This implies the existence and finiteness of the defining limits of the infinitesimal generator or rate matrix [Citation13, Chapter 8, Section 2.2]. Additional analytic properties of stochastic semigroups can be found at [Citation23, Chapter V].
Definition 5.3
The infinitesimal generator or rate matrix of a CTMC is with
equivalently, in matrix notation, componentwise,
Remark 5.2
Constants qij are transition rates from state i to state j and satisfy: qij ≥ 0 if i ≠ j, and
; i.e. Q has nonnegative off-diagonal entries and zero row sums (Q1 = 0, where 1 = (1, …, 1)T and 0 = (0, …, 0)T ), i.e. Q is a Markov generator.
Standard (finite state) stochastic semigroups {P(t)}t≥0 satisfy Backward and Forward Kolmogorov differential equations:
with initial condition P(0) = In, and unique solution
. We refer to [Citation24, Remark VII.2.3] for a digression about dynamics underlying above differential equations.
6. Continuous-time Markov evolution algebras
Let be a finite-dimensional real vector space with basis
and consider the continuous index set
. We denote by
the family of evolution algebras with multiplication:
and by
the structure matrix of
, for all t ≥ 0. Multiplication will be denoted simply by juxtaposition when t-value (time) is clear.
Definition 6.1
A continuous time Markov evolution algebra (CT-Markov EA) is a family
of evolution algebras whose structure matrices {A(t)}t≥0 w.r.t. (the natural basis)
form a standard stochastic semigroup.
We call rate matrix of a CT-Markov EA
to the matrix Q such that A(t) = etQ, t ≥ 0.
The existence of the rate matrix Q was discussed in the previous section. The following facts are stated as a lemma for further use. Proofs are straightforward.
Lemma 6.2
Let be a CT-Markov EA.
is a Markov evolution algebra, for all t ≥ 0, with M-natural basis
.
is the (nonzero) trivial Markov evolution algebra with structure matrix A(0) = In.
Given a Markov generator Q, the CT-Markov EA
(its structure matrices) is the unique solution of (backward and forward) Kolmogorov differential equations with initial condition A(0) = In. In fact, recall Q is the derivative of t → A(t) at origin.
In what follows, will denote a CT-Markov EA with standard stochastic semigroup {A(t)}t≥0 w.r.t. (a M-natural basis)
. For convenience, elements of
will be identified to the finite set of states Λ = {1, …, n} of the underlying continuous-time Markov chain.
Definition 6.3
The embedded Markov evolution algebra of a CT-Markov EA
is the Markov evolution algebra with structure matrix (w.r.t.
) the transition matrix
of the corresponding HDT embedded Markov chain, that is,
where
denotes the rate matrix of
.
Remark 6.1
is defined by the jump chain or embedded Markov chain
associated to the corresponding CTMC, where Yk = X(Wk), being
random variables defining the jump times or waiting times of the CTMC, that is, Wk is the time of the kth jump or time the CTMC remains in state X(Wk−1) before jumping into X(Wk) (and W0 = 0). Thus, since aij = P(Yk+1 = j| Yk = i), it holds qii = 0 if and only if aii = 1, i.e. if state i is absorbing with null change rate. Note
gathers probabilities
.
Example 6.4
Let us consider
Matrix Ae tells us that 1 → 2 → 3 → 4 → 1 [Citation12, Examples 5.3 and 5.8]. The solution of Kolmogorov differential equations gives us the CT-Markov evolution algebra
with multiplication defined by the standard stochastic semigroup:
Asymptotic behaviour of CT-Markov evolution algebras is given by their embedded Markov evolution algebras. Mimicking analogous notions for CT Markov chains, we introduce the following definitions.
Definition 6.5
Let , {A(t)}t≥0 and
as before, and consider
. Then ei leads to ej if aij(t) > 0 for some t ≥ 0 (we then write ei → ej), and elements ei and ej communicate (ei ↔ ej) if ei → ej and ej → ei.
We remark here that aij(t) > 0 for some t ≥ 0, implies aij(t) > 0 for all t > 0, and ei → ej in . Essential, inessential and communication classes of basic elements of CT-Markov evolution algebras are then defined similarly to the DT setting. This results into irreducibility of
to be equivalent to that of
.
Definition 6.6
A CT-Markov evolution algebra is basic simple if there exists no proper basic ideal common to
, for all t ≥ 0.
Theorem 6.7
A CT-Markov evolution algebra is basic simple if and only if Ae is irreducible.
Proof.
Let be a basic simple CT-Markov evolution algebra. Assuming Ae is reducible, there exist i, j ∈ Λ such that
, implying that for all
,
and therefore aij(t) = 0 for all t ≥ 0. Then
) is a basic ideal of
, for all t ≥ 0. Moreover, I is proper since
, contradicting the basic simplicity of
.
Assume now Ae is irreducible and (up to relabelling) a proper r-basic ideal of
for all t ≥ 0. Take i, j such that 1 ≤ i ≤ r < j ≤ n. Irreducibility of Ae implies
for some nonnegative integer k, and therefore aij(t) > 0 for some t ≥ 0, contradicting that I is a proper basic ideal of
.
It is then easy to prove that the CT-Markov evolution algebra in Example 6.4 is basic simple, whereas that appearing in (forthcoming) Remark 6.2 is not.
Proposition 6.8
Let be a CT-Markov EA with embedded Markov EA
.
(for all t ≥ 0) and
are nondegenerate.
is perfect for all t ≥ 0, but
is not necessarily perfect.
Proof.
(i) Follows from Proposition 3.2, and the first assertion of (ii) results then from det(A(t)) = det(etQ) = etr(tQ). Finally, we refer to [Citation12, Example 5.9] (see Remark 6.2 below) for a rate matrix Q defining a standard stochastic semigroup, hence a CT-Markov EA, with non-perfect embedded Markov EA, which by (i) above amounts to have det(Ae) = 0 as noted in Remark 3.1.
Remark 6.2
[Citation12, Example 5.9]
Consider the CT-Markov evolution algebra with structure matrix A(t) = etQ given by
and with corresponding rate matrix Q and Ae:
Clearly, state i = 4 is a source (with no loops at it) resulting in Ae containing a column of zeros. Therefore, det(Ae) = 0 implying that
is not perfect.
Proposition 6.8(ii) yields that properties of standard stochastic semigroups limit when a (discrete-time) Markov evolution algebra could exist within a CT-Markov evolution algebra (
, for some t ≥ 0), since any such Markov evolution algebra
needs to be perfect. This leads us to consider in the next section the embedding problem.
7. Embeddability of Markov evolution algebras
We will say that a Markov evolution algebra is embeddable into a CT-Markov EA
if its structure matrix A is embeddable, i.e. if there exists a rate matrix Q such that A = eQ.
If a Markov evolution algebra is embeddable, then {A(t) = etQ, t ≥ 0} is a standard stochastic semigroup defining a CT-Markov EA
, such that
, resulting from A = A(1). Uniqueness of the Markov generator Q is not, however, ensured, a question known as the identifiability problem (see [Citation8,Citation14,Citation25] and references therein).
We recall here the following results for further use.
Proposition 7.1
[Citation14, Proposition 2.1]
If a Markov matrix P is embeddable, P = eQ, with Markov generator Q, then:
σ(P) = eσ(Q), where σ(A) denotes the spectrum of a matrix A.
0 < det(P) ≤ 1, so
and det(P) = 1 only for P = In.
P is either reducible, or positive and hence primitive.
If pij > 0 and pjk > 0, then pik > 0.
Proposition 7.1 gives a first criterion to discard Markov evolution algebras not realizable by standard stochastic semigroups (i.e. non-embeddable).
Example 7.2
Consider the Markov evolution algebras appearing in Example 2.4(iv). The structure matrices of
and
are singular and that of
irreducible but not positive. Hence, none of these Markov evolution algebras is embeddable. Markov evolution algebra
(i.e.
) has structure matrix I2, so it is trivially embeddable since I2 = eQ for
.
The Markov evolution algebra
given by the random walk on the cycle graph with five vertices C5 (Example 2.5) is not embeddable, as Proposition 7.1(iv) is not satisfied. Indeed, notice
has structure matrix
with a12, a23 > 0 but a13 = 0.
Markov evolution algebras arising from birth and death processes on {0, 1…, N} with absorbing states 0 and N are not embeddable, as their structure matrices are singular (see Section 4.1).
Jukes–Cantor model (Section 4.3) is given by a 4 × 4 circulant matrix (a matrix is circulant when their rows arise from shifting them cyclically one position to the right). Embeddability of d × d circulant matrices (d ≥ 4) is characterized in [Citation14, Theorem 5.6]. The stochastic semigroup and rate matrix for Jukes–Cantor model can be found in [Citation24, 7.8.1]. The JC model is the simplest nucleotide model as it assumes all mutations occur with an equal rate. More complex models are also given in [Citation24, 7.8.1].
Remark 7.1
Despite being closely related to Markov processes and satisfying Chapman–Kolmogorov type conditions, CEAS are not necessarily embeddable. Consider for instance
and assume f(t) ∈ [0, 1] for all t ≥ 0 [Citation26, p. 352]. Then
is Markov, but singular, hence not embeddable.
As noticed above, studying CT-Markov evolution algebras becomes to be related to the embedding problem of Markov matrices. Despite of partial results for small dimensions, some of the most recent and general results on this problem can be found in [Citation25].
To conclude, and despite similar results on the embeddability of Markov evolution algebras that could be derived from characterizations given, for instance, in [Citation14,Citation25], we consider the case of 2-dimensional Markov evolution algebras. For this simple case, the embeddability of 2-dimensional Markov evolution algebras are fully determined by the following results on 2 × 2 Markov matrices, being moreover the Markov generator unique.
Proposition 7.3
[Citation14, Theorem 3.1, Corollary 3.3]
Let with a, b ∈ [0, 1]. Then, the following statements are equivalent:
P is embeddable.
0 < det(P) = 1 − (a + b) ≤ 1.
1 < tr(P) ≤ 2, i.e. 0 ≤ a + b < 1.
P has positive spectrum σ(P) = {1, 1 − (a + b)} (positive eigenvalues only).
Moreover, if the above equivalent conditions hold, there exists a unique rate matrix Q such that P = eQ. More precisely,
Corollary 7.4
If is embeddable, then a, b ∈ [0, 1).
Proof.
It suffices to note that a = 1 would imply det(P) = −b < 0 contradicting Proposition 7.3(ii) (similarly det(P) = −a < 0 if b = 1).
Theorem 7.5
Let be a 2-dimensional Markov evolution algebra. Then,
is embeddable into a CT-Markov evolution algebra
if and only if:
is perfect and has two 1-basic ideals. (More precisely,
is the trivial evolution algebra with structure matrix A = I2.)
is perfect and has a 1-basic ideal.
is perfect and (basic) simple.
Then the rate matrix Q of is unique, and as described in Proposition 7.3.
Proof.
Let be a 2-dimensional Markov evolution algebra with structure matrix
. We first assume
is embeddable, so that by Corollary 7.4 we necessarily have a, b ∈ [0, 1). Then, by Proposition 7.3:
If a = b = 0, i.e. if A = I2, then
is a non-simple trivial evolution algebra with 1-basic ideals
, i = 1, 2.
If a = 0 and b ≠ 0 (thus det(A) = 1 − b ≠ 0 ), then
, is a 1-basic ideal of
. (Analogously if a ≠ 0 and b = 0, considering then
.)
If ab ≠ 0 (thus det(A) = 1 − a − b ≠ 0 ), then
is perfect and (basic) simple [Citation11, Proposition 2.7].
Conversely, the structure matrix of any of the 2-dimensional Markov evolution algebras listed above is embeddable by Proposition 7.3.
7.1. Further comments
Tian's target of studying continuous evolution algebras, even under the additional assumptions considered here, has been proved to be connected to the classical problem on the embeddability of Markov matrices [Citation8]. Current research on this topic, see for instance [Citation25], would contribute to better understand continuous-time evolution algebras. Some more problems are still to be considered, as for instance, the representation of (Markov) evolution algebras as solutions of differential equations (see Lemma 6.2(iii)) or the case of evolution algebras with complex structure matrices with row sums equal to one.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Tian JP. Evolution algebras and their applications. Berlin: Springer; 2008. (Lecture notes in mathematics; 1921).
- Tian JP, Vojtechovsky P. Mathematical concepts of evolution algebras in non-Mendelian genetics. Quasigroups Relat Syst. 2006;14(1):111–122.
- Cabrera Y, Molina MS, Velasco MV. Evolution algebras of arbitrary dimension and their decompositions. Linear Algebra Appl. 2016;495:122–162.
- Camacho ML, Gómez JR, Omirov BA, et al. The derivations of some evolution algebras. Linear Multilinear Algebra. 2013;61(3):309–322.
- Casas JM, Ladra M, Omirov BA, et al. On evolution algebras. Algebra Colloq. 2014;21(2):331–342.
- Elduque A, Labra A. Evolution algebras and graphs. J Algebra Appl. 2015;14(7):1550103. 10pp.
- Casas JM, Ladra M, Omirov BA, et al. A chain of evolution algebras. Linear Algebra Appl. 2011;435:852–870.
- Davies EB. Embeddable Markov matrices. Electron J Probab. 2010;15:1474–1486.
- Mellon P, Velasco MV. Analytic aspects of evolution algebras. Banach J Math Anal. 2019;13(1):113–132.
- Seneta E. Non-negative matrices and Markov chains. Revised reprint of the second (1981) edition. New York (NY): Springer; 2006. (Springer series in statistics).
- Cabrera Y, Kanuni M, Molina MS. Basic ideals in evolution algebras. Linear Algebra Appl. 2019;570:148–180.
- Allen LJS. An introduction to stochastic processes with applications to biology, Second edition. Boca Raton (FL): CRC Press; 2011. 2010.
- Brémaud P. Markov chains. Gibbs fields, Monte Carlo simulation, and queues. New York (NY): Springer-Verlag; 1999. (Texts in applied mathematics; 31).
- Baake M, Sumner J. Notes on Markov emdedding. Linear Algebra Appl. 2020;594:262–299.
- Casanellas M, Fernandez-Sanchez J, Roca-Lacostena J. Embeddability and rate identifiability of Kimura 2-parameter matrices. J Math Biol. 2020;80:995–1019.
- Khudoyberdiyev AK, Omirov BA, Qaralleh I. Few remarks on evolution algebras. J Algebra Appl. 2015;14(4):16pp.
- Narkuziyev BA. Some remarks on evolution algebras corresponding to permutations. Preprint arXiv:2005.05793.
- Murodov ShN. Classification of two-dimensional real evolution algebras and dynamics of some two-dimensional chains of evolution algebras. Uzbek Mat Z. 2014;2:102–111.
- Cadavid P, Montoya MLR, Rodríguez PM. On the isomorphism between evolution algebras of graphs and random walks. Preprint arXiv:1710.10516v2.
- Cadavid P, Montoya MLR, Rodríguez PM. The connection between evolution algebras, random walks and graphs. J Algebra Appl. 2020;19:2050023. 28pp.
- Daley DJ, Gani J. Epidemic modelling: an introduction. Cambridge: Cambridge University Press; 1999. (Cambridge studies in mathematical biology; 15).
- Casanellas M, Kedzierska AM. Generating Markov evolutionary matrices for a given branch length. Linear Algebra Appl. 2013;438:2484–2499.
- Freedman D. Markov chains. New York (NY): Springer Verlag; 1983. Corrected reprint of the 1971 original.
- Pardoux E. Markov processes and applications: algorithms, networks, genome and finance. Paris: Wiley; 2008. (Wiley series in probability and statistics).
- Casanellas M, Fernandez-Sanchez J, Roca-Lacostena J. The embedding problem for Markov matrices. Preprint arXiv:2005.00818v1.
- Rozikov UA, Murodov ShN. Dynamics of two-dimensional evolution algebras. Lobachev J Math. 2013;34(4):344–358.
