Abstract
The regulation of brain cytochrome P450 enzymes (CYPs) is different compared with respective hepatic enzymes. This may result from anatomical bases and physiological functions of the two organs. The brain is composed of a variety of functional structures built of different interconnected cell types endowed with specific receptors that receive various neuronal signals from other brain regions. Those signals activate transcription factors or alter functioning of enzyme proteins. Moreover, the blood-brain barrier (BBB) does not allow free penetration of all substances from the periphery into the brain. Differences in neurotransmitter signaling, availability to endogenous and exogenous active substances, and levels of transcription factors between neuronal and hepatic cells lead to differentiated expression and susceptibility to the regulation of CYP genes in the brain and liver. Herein, we briefly describe the CYP enzymes of CYP1-3 families, their distribution in the brain, and discuss brain-specific regulation of CYP genes. In parallel, a comparison to liver CYP regulation is presented. CYP enzymes play an essential role in maintaining the levels of bioactive molecules within normal ranges. These enzymes modulate the metabolism of endogenous neurochemicals, such as neurosteroids, dopamine, serotonin, melatonin, anandamide, and exogenous substances, including psychotropics, drugs of abuse, neurotoxins, and carcinogens. The role of these enzymes is not restricted to xenobiotic-induced neurotoxicity, but they are also involved in brain physiology. Therefore, it is crucial to recognize the function and regulation of CYP enzymes in the brain to build a foundation for future medicine and neuroprotection and for personalized treatment of brain diseases.
Introduction
Studies of the last years indicate that the regulation of brain cytochrome P450 enzymes (CYPs) is different compared with respective hepatic enzymes. This may result from anatomical bases and physiological functions of the two organs. The brain is a heterogeneous organ, consisting of various functional structures that contain different cell types receiving extensive neuronal inputs from other regions of the brain. For instance, noradrenergic, dopaminergic and serotonergic innervations come to the forebrain from the brain stem, while glutamatergic neurons of the cortex, hippocampus or amygdala send their projections to other brain areas (Stahl Citation2013). Inputs that reach the cell are transmitted into the neuronal cell interior via specific membrane or intracellular receptors and corresponding signal transduction pathways, to activate transcription factors or to alter protein functioning (e.g. enzyme activity). Each brain structure receives unique innervations, and each cell type possesses its own pre- and postsynaptic receptors that mediate interneuronal and interstructural connections. Therefore, the physiological regulation of particular genes differs between brain regions. These differences in the regulatory modes concern also the genes encoding cytochrome 450 enzymes which are expressed in the brain, though in much lower levels than in the liver.
Moreover, the specific anatomic construction of the blood-brain barrier (BBB), endowed with an efflux transport system, does not allow free penetration of all endogenously produced substances or xenobiotics from the periphery into the brain, except for some BBB-free areas localized around cerebral ventricles. The above-mentioned distinct innervations, receptor-transduction pathways, and availability of endogenous and exogenous compounds may be a reason for inter-regional differences in the regulation of CYP gene expression and enzyme activity in the brain, as well as for inter-organ differences in the regulation of those enzymes between the brain and the liver.
For example, some hormones excreted into the blood (e.g. growth hormone) or drugs/drug metabolites do not easily penetrate the BBB (Lai et al. Citation1991; Nyberg and Burman Citation1996), and there is an essential difference in levels of CYP gene-regulating receptors (e.g. CAR, PXR, AhR, PPAR, RXR, GHR) between the brain and liver (Lobie et al. Citation1993; Nyberg and Burman Citation1996; Petersen et al. Citation2000; Nishimura et al. Citation2004). Therefore, it seems that differences in neurotransmitter signaling, availability to endogenous and exogenous active substances, and levels of transcription factors between neuronal and hepatic cells lead to differentiated expression and susceptibility to the regulation of CYP genes in the brain (brain areas) and liver.
Brain CYP enzymes exert a direct impact on a wide range of processes, with confirmed effects on behavior, neurotoxicity, and drug response (e.g. analgesics, anesthetics, antidepressants, antipsychotics) (Toselli et al. Citation2016; McMillan and Tyndale Citation2018), playing a potential role in the metabolism of important endogenous molecules, drugs, and neurotoxins in the brain. Local brain metabolism of endogenous substrates and drugs that cross the BBB may contribute to inter-individual variability in therapeutic responses. This may be implicated in human behavioral reactions, as well as in stress, depression, schizophrenia, cognitive processes, learning, and memory (Haduch et al. Citation2018; Stingl Citation2018).
This article briefly describes the catalytic competence and distribution of CYP1-3 families in the brain and discusses brain-specific regulation of CYP genes compared to the regulation of liver CYP genes.
General characteristics and origin of cytochrome P450s
Cytochrome P450s are a superfamily of enzymes ubiquitously expressed in all kingdoms of life: animals, insects, plants, fungi, protists, bacteria, archaea, and viruses. Currently, more than 52,675 CYP sequences have been documented with a variety of critical physiological roles. Humans are regularly exposed to diverse chemicals, drugs, food additives, and pollutants that could eventually impair cellular stability and disrupt metabolism with damaging health effects. The unique defense systems that contain hemeprotein, termed cytochrome P450 enzymes, have been known since the 1960s (Raw et al. Citation1960; Omura and Sato Citation1962), and the number of P450s classified in the superfamily has grown to more than 21,000 members by August 2013 (Cytochrome P450 Stats. (2013) https://drnelson.uthsc.edu/P450.stats.Aug2013.png (accessed 20 January 2020). Naming of these enzymes is based on their spectral absorption wavelength. The spectrum of the reduced form of the enzymes exhibits a Soret peak at 450 nm when the enzyme is saturated with carbon monoxide, reflecting the retention of cysteine thiolate proximal coordination in this form of the enzyme (Omura and Sato Citation1962; to Citation1964). In mammals, CYP enzymes are engaged in the biosynthesis and metabolism of steroid hormones (Ryan and Engel Citation1957; Estabrook et al. Citation1963), various lipid biofactors (Das et al. Citation1968; Lu and Coon Citation1968), including eicosanoids (Chacos et al. Citation1983; Iliff et al. Citation2007), vitamin D3 (Bhattacharyya and DeLuca Citation1973; ca Citation1974; Fukushima et al. Citation1978) and retinoids (Swindell et al. Citation1999), and can catalyze alternative pathways of neurotransmitter synthesis, especially dopamine (Hiroi et al. Citation1998) and serotonin (Yu et al. Citation2003).
Diverse CYP enzymes are found in different tissue and cell types co-occur in several separate subcellular compartments simultaneously. Most cytochrome enzymes are bound to membranes, such as the outer nuclear membrane, endoplasmic reticulum, mitochondria, Golgi, peroxisomes, and plasma membrane (Khandwala and Kasper Citation1973; Sagara et al. Citation1978; Williams et al. Citation2000). Soluble forms also exist, but rather in yeast and prokaryotes. The highest quantity of CYP1A1 protein occurs in the brain tissue and was identified in the cytosolic fraction (Meyer et al. Citation2002).
It has been proposed that CYPs originated from the prokaryotic sterol 14α-demethylase CYP51, which is regarded as an ancient cytochrome P450 (Yoshida et al. Citation1997; Parvez et al. Citation2016). Primordial CYP gene products in vertebrates perhaps first evolved to fulfill vital life functions, then handling plant substrate degradation and drug metabolism abilities. Although the CYP2G and CYP2T subfamilies encode functional genes in rodents, they only seem to encode pseudogenes in humans, suggesting that whatever functions these genes had approximately 80 million years ago (at the time of the mammalian radiation), they are no longer needed in humans. Mice deficient in the Cyp2e1 gene seem to be outwardly healthy, but are very resistant to benzene’s toxic effects, indicating a role in xenobiotic metabolism for this subfamily (Gonzalez and Kimura Citation2001; Nebert and Russell Citation2002).
Examination of the genomes of divergent organisms has revealed that many of the CYP genes are present in accumulated clusters on chromosomes (Doddapaneni et al. Citation2005). Such clustering presents strong evidence that these genes originated via gene duplication, either as discrete events or as a part of whole-genome duplications (Kumar and Hedges Citation1998; Finta and Zaphiropoulos Citation2000; Turman et al. Citation2006; Feyereisen Citation2011).
Structure
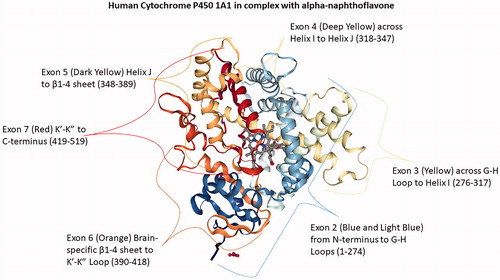
Structures of all of the cytochrome P450s share certain elements and show several similarities. These proteins are organized into a similar series of helices and folds. The folds are predominantly α-helical, with a small proportion of β-sheets. Its twelve helices are designated A to L and are named in alphabetical order starting from the N-terminus (Poulos et al. Citation1987). Substrate recognition sequences, the most critical structural regions of CYPs (Gotoh Citation1992; Yasukochi and Satta Citation2015; Tietz et al. Citation2017), include a highly conserved I-helix bearing a threonine amino acid, which plays an essential role in catalytic processes, the F/G-loop, and the F and G-helices, together with the B/C-loop, which form a ‘lid’ over the active site cavity (Otyepka et al. Citation2007). The active site is deeply embedded within the structure and is coupled to the outer region through a complex network of access and exit channels (Wade et al. Citation2004; Cojocaru et al. Citation2007). It houses a protoporphyrin IX (heme b) cofactor, which is connected to the CYP catalytic domain by a Fe-S bond to the thiolate group of a highly conserved cysteine (Mestres Citation2004; Šrejber et al. Citation2018). Interestingly, structural analysis of the CYP1A1 gene reveals an arrangement on Exon 6 that appears to be the protein domain responsible for activating cerebral activity of the enzyme () (Kommaddi et al. Citation2007; Annalora et al. Citation2017). Furthermore, alternative splicing might result in multiple transcript variants encoding distinct CYP enzymes (Zhuge and Yu Citation2004; Ren et al. Citation2005; Annalora et al. Citation2017).
Figure 1. Human Cytochrome P450 1A1 in complex with alpha-naphthoflavone. Exon 2 (1-275) rainbow blue extends from the N-terminus to G-H Loop; Exon 3 (276-317) G-H Loop to Helix I; Exon 4 (318-347) Helix I to Helix J; Exon 5 (348- 389) Helix J to β1-4 sheet, Exon 6 (390-418) Brain-specific β1-4 sheet to K’-K” Loop; and Exon 7 (419-512) K’-K” to C-terminus red. PDB ID: 4I8V (Walsh et al. Citation2013; Annalora et al. Citation2017; Rose et al. Citation2018).
In humans, 57 genes encoding various cytochrome P450 enzymes have been identified. These genes are classified into 18 families and 57 subfamilies based on their amino acid sequence similarity (Nelson Citation2011). Members of CYP 1, 2, and 3 families are primarily involved in xenobiotic metabolism and physiological processes in the brain, whereas other families are employed in metabolism of endogenous substrates.
Distribution, function, and regulation of brain cytochrome P450 − overview
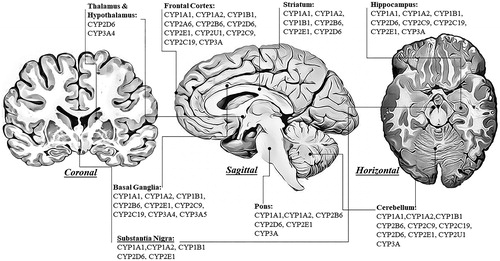
Xenobiotic metabolism principally occurs in the liver, which also exhibits the highest expression and concentration of transmembrane cytochrome P450 proteins. Total CYP protein content in the brain is significantly lower compared to the liver and is estimated to be 0.5%–2% of hepatic CYP content (Hedlund et al. Citation2001). The subcellular distribution of CYP1-3 family enzyme expression (mRNAs, proteins, activities) within the human brain has been thoroughly described in a comprehensive review by Toselli et al. (Citation2016). Expression levels and activities of particular CYP enzymes vary between brain structures (), and enzyme proteins are usually present in both microsomal and mitochondrial fractions. Although total CYP protein content in the brain is significantly lower than in the liver, in some cell types, it reaches levels similar to that observed in the liver (Miksys and Tyndale Citation2013).
Figure 2. The expression of human cytochrome P450 enzymes involved in the metabolism of xenobiotics in the selected brain regions shown in coronal, sagittal, and horizontal brain sections.
A few CYPs are primarily expressed in the brain. These include CYP7B, explicitly existing in the hippocampus of rat and mouse (Stapleton Citation1995), CYP26B, expressed in the cerebellum and pons (White et al. Citation2000), and CYP46, displaying widespread expression in the brain (Lund et al. Citation1999). These three brain-specific CYP enzymes are not involved in the metabolism of foreign compounds. Still, these enzymes are essential for endogenous functions, such as regulation of neurosteroids, cholesterol, and vitamin A metabolism (Lin and Smith Citation1974; Björkhem et al. Citation1997; Lund et al. Citation1999; Sakai et al. Citation2001).
Additionally, the brain is protected by the vigorous neurovascular defense of the BBB and the blood-cerebrospinal fluid barrier against unwanted compounds with selective entry restricted by ATP‐binding cassette (ABC) efflux transporters (Dauchy et al. Citation2008). CYP1B1 is expressed at the human BBB, possibly acting in conjunction with transport proteins to regulate xenobiotic penetration (Dauchy et al. Citation2008; Dutheil et al. Citation2009). In addition to CYP1B1 transcripts, CYP2U1 was also detected in human brain microvessels (Shawahna et al. Citation2011). The highest expression of CYP2B was observed in the dentate gyrus of the hippocampus and astrocytes adjacent to cerebral blood vessels, possibly playing a similar role to CYP1B1 in neuroprotection at the BBB by cooperating with transporters (Miksys et al. Citation2003; McMillan and Tyndale Citation2018). The BBB expresses high levels of different CYP enzymes, forming a metabolic barrier that regulates blood flow, compound flow, and signal transmission during inflammation (Meyer et al. Citation2007).
All cytochrome P450 enzymes exhibit characteristic alternative splicing that results in multiple transcript variants encoding distinct CYP enzymes. These enzymes are primarily expressed extrahepatically. Evidence for CYP1B1 and CYP2U1 expression in brain microvessels of the BBB has been revealed by proteomics analysis (Shawahna et al. Citation2011).
In neurons, cytochrome P450 has other functions: in regions such as the hypothalamus (Tsuruo et al. Citation1994; Abdelgadir et al. Citation1997), the hippocampus (Rosenbrock et al. Citation1999; Hagemeyer et al. Citation2003; Hojo et al. Citation2004; Meyer et al. Citation2006), and the striatum (Schilter and Omiecinski Citation1993; Huang et al. Citation2000), it provides signaling molecules (steroids and fatty acids) necessary for neuronal formation and development. CYP enzymes are differentially expressed in individual regions of the brain and in specific neuronal and glial cells (). The highest CYP content is found in the brain stem (the structure containing cell bodies of dopamine, noradrenaline, and serotonin neurons), cerebellum (Purkinje cells in particular; the structure engaged in motor control, cognitive and emotional functions), hippocampus (the structure involved in learning and memory), ventral striatum containing the nucleus accumbens (the structure engaged in cognitive functions, reward, and addiction), and dorsal striatum (the structure involved in motor functions) (Dutheil et al. Citation2008). Drug metabolizing CYPs, 1A1, 1A2, 2A6, 2B1, 2E1, and 3A4, occur predominantly in neurons (Bhagwat et al. Citation2000; Singh et al. Citation2012), whereas CYP1B1 and CYP2D6 are expressed in both neurons and glial cells (Gilham et al. Citation1997; Muskhelishvili et al. Citation2001; Miksys et al. Citation2003).
Cerebral CYP enzymes may generate a microenvironment of various endogenous substrates, drugs, and their metabolites, which cannot be monitored in the blood (Miksys and Tyndale Citation2004). Many CYPs have currently been detected in the brain. Although in the brain expression of CYP1-3 enzymes is generally lower than in the liver, these enzymes may regulate local levels of substances in particular regions and cell types of the brain (Norman and Neal Citation1976; Sasame et al. Citation1977; Warner et al. Citation1988; Miksys and Tyndale Citation2013; Toselli et al. Citation2016). It should be taken into consideration that due to analytical limitations, detection of CYP activity ex vivo is still restricted due to low enzyme levels but also due to rapid degradation. Notably, nearly 50% of brain CYP2D activity is lost within a week of freezing (Tyndale et al. Citation1999). There are also practical complications resulting from the use of antibodies raised against hepatic CYP forms for enzymes found in the nervous system since they can differ in immunoreactivity (Miksys et al. Citation2002). These difficulties in performing research on the regulation of brain cytochrome P450 expression and function (activity) deserve a great deal of consideration as there is not always a good correlation between mRNA, protein, and enzyme activity. Another issue to consider is that altered cell signaling (e.g. cAMP production) in response to neuroactive drugs may affect the phosphorylation processes and, in turn, enzyme protein synthesis, function, or degradation (Oesch-Bartlomowicz and Oesch Citation2003, Citation2005). Furthermore, exposure to environmental xenobiotics or treatment with drugs producing reactive metabolites (e.g. some neuroleptics and antidepressants) may lead to interference with protein synthesis or binding of those metabolites to CYP proteins, affecting enzymatic activity in this way (discussed in Daniel Citation2005).
CYP enzyme induction is mostly transcriptional, even though non-transcriptional mechanisms, such as stabilization of mRNA, enzyme stabilization, or inhibition of the protein degradation pathway, have also been reported. Noncoding RNAs, as microRNAs (miRNAs), are involved in the posttranscriptional regulatory mechanisms of major drug-metabolizing enzymes (Dluzen and Lazarus Citation2015). miRNAs are non-coding RNAs with a span of approximately 22 nucleotides and play a negative regulatory role by binding to the 3′- or 5′-UTR of target mRNA to inhibit translation or initiate mRNA degradation. All these mechanisms of CYP enzyme regulation take place in the brain, though in a tissue-specific manner, often differing for a particular CYP enzyme compared to the liver.
The CYP1 family
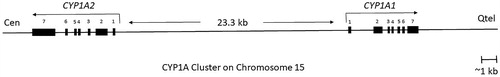
The CYP1 family consists of two subfamilies, encompassing three functional genes, CYP1A1, 1A2, and 1B1, in humans. CYP1A1 and CYP1B1 are highly conserved among species (Martignoni et al. Citation2006). Two related family members, the CYP1A1 and CYP1A2 genes, which consist of seven exons and six introns, are located approximately 23.3 kb away from each other on chromosome 15 (), whereas CYP1B1 consists of only three exons and is located on chromosome 2.
Figure 3. An overview of human CYP1A cluster, located on chromosome 15q22-qter with CYP1A1 reading toward the telomere and CYP1A2 reading toward the centromere (Corchero et al. Citation2001). The distance between the start of transcription of CYP1A1 and CYP1A2 is 23.3 kb. There are 13 XRE (xenobiotic‐responsive element) at the core DNA sequences that may participate in CYP1A induction following activation of the Ah receptor (adapted from Galijatovic et al. Citation2004; Ye et al. Citation2019).
CYP1A-catalyzed reactions include hydroxylation of aromatic compounds. Specifically, CYP1A1 is primarily engaged in the metabolism of aromatic hydrocarbons, while CYP1A2 transforms aromatic amines and heterocyclic derivatives. Constitutive expression of CYP1A1 was found in both human and rat brain (McFadyen et al. Citation1998; Chinta et al. Citation2005). CYP1A1 mRNA and protein are localized predominantly in neurons of the cerebral cortex, Purkinje and granule cell layers of the cerebellum, and pyramidal neurons of the hippocampus (in CA1, CA2, and CA3 subfields). Moreover, the presence of a splice variant was demonstrated in the human brain but not in the liver from the same individual (Chinta et al. Citation2005). Specific CYP1A1 and CYP1A2 activity of 7-ethoxycoumarin O-deethylase has been observed in mitochondria from several human brain regions, showing the highest levels in the striatum and pons and the lowest levels in the cortex and cerebellum (Bhagwat et al. Citation2000).
CYP1A is a central enzyme during phase I drug metabolism and contributes to the biotransformation of nearly 9% of clinical drugs, such as caffeine, analgesics, antipyretics, antipsychotics, antidepressants, and anti-inflammatory drugs (Lu et al. Citation2020). In addition to medicines, CYP1A is also involved in the biological activation or deactivation of a large number of pollutants in the environment, such as benzopyrene, aristolochic acid I, ellipticine, PhIP (2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine), and IQ (2-amino-3-methylimidazo[4,5-f]quinolone) (Abdel-Razzak et al. Citation1994; Josephy et al. Citation2000; Stiborová et al. Citation2001; Cheung et al. Citation2005; Kotrbová et al. Citation2011). Furthermore, CYP1A1 and CYP1A2 metabolize sex hormones (including progestogens, androgens, and estrogens), retinol, melatonin, linoleic acid, phosphatidylcholine, and uroporphyrinogen (Shou et al. Citation1997; Niwa et al. Citation1998; Sinclair et al. Citation1998; Yamazaki et al. Citation1998; Yun et al. Citation1999; Chen et al. Citation2000; Moran et al. Citation2000; Ohe et al. Citation2000; Schwarz et al. Citation2000; Von Bahr et al. Citation2000; Ma et al. Citation2005; Phillips et al. Citation2011). Progesterone plays a crucial role in the physiological function of the brain by producing other endogenous steroids, neurosteroids (Baulieu and Schumacher Citation2000).
CYP1A catalyzes the synthesis and metabolism of melatonin via 5-hydroxylation. Melatonin is the primary hormone secreted by the pineal gland (Hardeland Citation2010; Tordjman et al. Citation2017). Circadian production of melatonin by the pineal gland explains its chronobiotic influence on the organism activity, including endocrine and non-endocrine rhythms. Other roles of melatonin, including its antioxidant and anti-inflammatory properties, its genomic effects, and its capacity to modulate mitochondrial homeostasis, are linked to the redox status of cells and tissues (Acuña-Castroviejo et al. Citation2014). Melatonin, a metabolite in the tryptophan pathway, may also be involved in human affective disorders (Srinivasan et al. Citation2006; Lu et al. Citation2020).
CYP1B1 is primarily localized in the putamen (part of the dorsal striatum) and the temporal and frontal lobe (McFadyen et al. Citation1998; Rieder et al. Citation1998). CYP1B1 is found in both the mitochondrial and microsomal fractions of astrocytes in the human frontal lobe, hippocampus, substantia nigra, and cerebellum (Shawahna et al. Citation2011).
Regulation
In humans, transcriptional activation of the CYP1A1 and CYP1A2 genes is controlled simultaneously by a dual promoter construct existing between these two genes and acting bidirectionally. The promoter contains arylhydrocarbon receptor (AhR) response elements (Ueda et al. Citation2006; Jorge-Nebert et al. Citation2010). Expression of nuclear AhR may be higher in a particular brain area than in the liver. For example, the rat olfactory cortex, hippocampus, and hypothalamus revealed the highest expression compared to low levels in the amygdala (Petersen et al. Citation2000). Both CYP1A genes are highly inducible by various xenobiotics, which act as AhR ligands, such as methylcholanthrene, β-naphthoflavone, polycyclic aromatic hydrocarbons, dioxins, and other compounds (Nebert et al. Citation2004). There is evidence of the role of nicotine in the induction of CYP1A1 and CYP1A2 enzymes in vitro in the brains of mice and rats (Anandatheerthavarada et al. Citation1993; Singh et al. Citation2008).
AhR is also engaged in the regulation of the CYP1B1 gene (Shehin et al. Citation2000; Chiaro et al. Citation2007). AhR stimulation initiates the synthesis of cytochrome P450 family 1 enzymes (CYP1s), which oxidize AhR ligands, leading to their metabolic clearance and detoxification (Schiering et al. Citation2017). The CYP1 enzymes have a critical feedback role that limits AhR signaling (Chiaro et al. Citation2007). Expression of CYP1A1 is controlled by negative feedback from AhR in enteric neurons (Obata et al. Citation2020). Serotonin (5-HT) induces CYP1A1 mRNA expression and CYP1A1 activity in intestinal epithelial cells (Manzella et al. Citation2018). Interestingly, 5-HT is a CYP1A1 substrate that competes with AhR ligands for CYP1A1 metabolism, preventing ligand deactivation and stimulating sustained AhR signaling (Manzella et al. Citation2020).
Covalent methylation of nuclear chromatin in the brain plays unique and essential roles in the nervous system (Mo et al. Citation2015). Interestingly, the CYP1A1 and CYP1B1 gene promoters are hypermethylated in hepatocytes. The transcript levels of CYP1A1 and CYP1B1 were 10-fold higher after combinatorial inhibition of DNA methyltransferases (DNMTs) and histone deacetylases (HDACs). Incomplete demethylation after DNMT inhibition is likely to be associated with enriched H3K27me3 in a regulatory region of CYP1A2 (Park et al. Citation2015). H3K27me3 plays a role in silencing genes for a short time or persistently and concerns neuron subtype-specific transcription factors (Hon et al. Citation2012). Furthermore, selective and transient silencing of CYP1A1, but not CYP1B1, was detected in human hepatoma cell lines. The endogenous ligand of the AhR 6-formylindolo[3,2-b]carbazole (FICZ) transiently represses CYP1A1 by targeting DNA (cytosine-5)-methyltransferase 3 A (DNMT3A) and is affiliated with specific DNA methylation of the CYP1A1 gene promoter (Ghaedi et al. Citation2020). Interestingly, male and female cells from similar tissues have sex-distinct patterns of DNA methylation of CYP family members: Cyp1a1, Cyp2e1, and Cyp7b1 exhibit a differential magnitude of gene expression (Penaloza et al. Citation2014). The epigenetics of sex differences in the brain has been acknowledged (McCarthy et al. Citation2009), and it has been demonstrated that DNA methylation changes occur in response to environmental stress, with some of these changes occurring in a sex-specific manner (Sterrenburg et al. Citation2011; Essex et al. Citation2013).
Sex hormones regulate cerebral drug metabolism through monitoring the expression of specific forms of microRNAs (Li et al. Citation2015). Highly expressed in the brain, short non-coding RNA miR-27b (Chi et al. Citation2009) targets and negatively regulates CYP1B1 (Liu et al. Citation2020). Current evidence suggests that CYP1A2 and CYP2E1 activity is higher in males than in females, while CYP3A, one of the most clinically relevant CYP isoforms, appears to have higher activity in females (Scandlyn et al. Citation2008). CYP1A2 activity is higher in men than in women for several substrates, including the conversion of caffeine to paraxanthine (Relling et al. Citation1992; Ou‐Yang et al. Citation2000).
Recently, the mechanism of CYP1B1 regulation by glutamate (Yu et al. Citation2019) has been demonstrated. Glutamate is a primary amino acid neurotransmitter known for its role as an excitatory neurotransmitter in the mammalian nervous system. An in vitro study showed that CYP1B1 preferentially produced midchain hydroxyeicosatetraenoic fatty acids (HETEs), almost 55% of its total metabolites (Choudhary et al. Citation2004). Increases in CYP1B1 mRNA levels and binding of cAMP response element-binding protein (CREB) to CYP1B1 promoters following high glutamate treatment were observed. This effect was reduced by an mGlu5 receptor antagonist (Yu et al. Citation2019).
The CYP2 family
The CYP2 gene family represents approximately 30% of human liver CYP isoforms and is the most prominent cytochrome P450 family (Daly Citation2016). Eleven subfamilies represent that family in the human body and all share a pattern of gene sequence arrangements with nine encoding exons, except for CYP2U1 and CYP2R1, which have only five exons (Cheng et al. Citation2003; Dhers et al. Citation2017). CYP2 genes are located on different chromosomes and are organized in multi-gene clusters.
CYP2A subfamily
Three dominant gene clusters comprise the CYP2A-T cluster on chromosome 19q13.2, which contains two significant genes, CYP2A6 and CYP2B6 () (Hoffman et al. Citation2001). The CYP2A-T cluster diverged through duplications and inversions and comprises six genes and seven pseudogenes in humans (Wang et al. Citation2003). Of the six human genes, four encode xenobiotic-metabolizing CYPs, and two are orphans of unknown function (Sezutsu et al. Citation2013). The human CYP2A6 metabolic pathway is responsible for 80% of oxidative metabolism of the psychoactive tobacco ingredient nicotine to the inactive cotinine (Messina et al. Citation1997). CYP2A6 expression in the brain is relatively low compared to other CYPs (Human Protein Atlas) (Koskela et al. Citation1999; Uhlén et al. Citation2015), and its role therein has not yet been well recognized.
Figure 4. An overview of the CYP2A-T gene cluster on human chromosome 19. Arrows and blocks represent the position, relative length and direction of transcription for each locus (adapted from Hoffman et al. Citation2001), ‘‘P’’ at the end of a gene name indicates a pseudogene.

Regulation
Cytochrome P450 monooxygenases of the CYP2A subfamily play significant roles in xenobiotic processing in the liver and metabolic activation in extrahepatic tissues. Many CYP2A transcripts and enzymes are induced by xenobiotic compounds (Su and Ding Citation2004). For instance, human CYP2A6 is primarily expressed in the liver, but lower levels are found in tissues of the respiratory tract, including the nasal mucosa, trachea, and lung, as well as the skin (Janmohamed et al. Citation2001; Saeki et al. Citation2002). Interestingly, one of the CYP2A6 alleles (rs113288603) expressed in the cerebellar hemisphere is associated with hearing loss in elderly smokers (Polimanti et al. Citation2017).
The expression of CYP2A genes is regulated by a conserved transcription factor-binding site called the nasal predominant transcriptional activating (NPTA) element, which was identified in the proximal promoter region of the CYP2A3, Cyp2a5, and CYP2A6 genes (Zhang and Ding Citation1998). The NPTA element is related to the nuclear factor 1 (NF1) element (Zhang et al. Citation2000; Ling et al. Citation2004). The NF1 genes are a group of transcription elements that bind to CAATT-boxes and control the beginning of gliogenesis in the embryonic spinal cord and differentiation of astrocytes later in gliogenesis (Deneen et al. Citation2006; Molofsky et al. Citation2013). NF1A expression in astrocytomas is associated with improved survival (Song et al. Citation2010). Interestingly, neuronal expression of the 5-HT3 serotonin receptor gene requires NF1 complexes (Bedford et al. Citation1998).
A genetic polymorphism in the promoter region of the CYP2A6 gene modulates the expression levels of CYP2A6 mRNA and protein in humans (Yoshida et al. Citation2003; von Richter et al. Citation2004). Sex differences in gene expression and splicing are common in the adult human brain, being measurable in all major brain regions (Trabzuni et al. Citation2013). There is already evidence for such a discrepancy of CYP2A gene expression in the liver, where higher hepatic activity of CYP2A6 was observed in women (Sinues et al. Citation2008; Al Koudsi et al. Citation2010). Bilirubin level reduction was shorter in women than in men (Kao et al. Citation2017). Bilirubin treatments significantly promote CYP2A6 expression in hepatocyte progenitor cells (HepRG cells). Analysis showed similar levels of AhR (bilirubin responsive nuclear receptor) and estrogen-estrogen receptor α (ESR1) binding to the promoter region of CYP2A6 (Kao et al. Citation2017). ESR1 is highly expressed in the pituitary gland (The Human Protein Atlas), as well as in glutamatergic and GABAergic neurons, where it is vital for regular sexual maturity onset, estrogen feedback, and fertility in female mice (Cheong et al. Citation2015).
Mouse hepatic CYP2A4 and CYP2A5 (Lavery et al. Citation1999; Viitala et al. Citation2001) and rat CYP2A1 (Seng et al. Citation1996) are controlled by the circadian rhythm. Mouse enzymes present the highest expression levels in the evening through a mechanism that involves the albumin D-site-binding protein (DBP) transcription factor (Lavery et al. Citation1999). DBP is a transcriptional guiding protein that exhibits robust circadian rhythms in tissues with high clock gene expression, such as the suprachiasmatic nucleus of the hypothalamus and the liver (Gachon et al. Citation2004). It is still unknown whether similar patterns of CYP regulation are adopted by cerebral cells. Interference with circadian rhythms in humans may lead to various pathological conditions, including depression, metabolic syndrome, and cancer (reviewed in Takahashi et al. Citation2008; Sahar and Sassone-Corsi Citation2012). Human cerebral cells are involved in the fundamental function of awareness during daytime activities that is represented by intensive neural transmission of action potentials (Cook Citation2008). As a consequence, adult brain requires enhanced clearance of metabolites during sleep (Xie et al. Citation2013).
CYP2B subfamily
CYP2B is widely expressed within the brain (Gervot et al. Citation1999; Bhagwat et al. Citation2000; Rosenbrock et al. Citation2001; Miksys et al. Citation2003). The expression of CYP2B in the monkey brain is the highest in Purkinje cells in the cerebellum, pyramidal cells of the cortex, and neurons in the substantia nigra (the structure localized in the midbrain, containing primarily cell bodies of dopaminergic neurons) (Miksys et al. Citation2002). However, the expression of CYP2B in rats and mice is the highest in neurons of the cerebellum, hippocampus, and thalamus (Volk et al. Citation1991; Lee et al. Citation2006). CYP2B is involved in the local metabolism of steroids and drugs. Brain cytochrome P450 was shown to contribute to the biotransformation of neuroactive drugs and to affect both their local (but not plasma) concentrations and central pharmacological/toxicological responses (McMillan and Tyndale Citation2018). For example, brain CYP2B significantly contributes to the local metabolism and concentration of the anesthetic propofol, which correlates with sleep time in the rat (Khokhar and Tyndale Citation2011). Brain CYP2B influences also nicotine levels in the brain (but not in plasma) and its behavioral effects in the self-administration test (Garcia et al. Citation2015).
Regulation
As mentioned before, the CYP2A-T cluster on chromosome 19q13.2 contains the CYP2A6 and CYP2B6 genes () (Hoffman et al. Citation2001). The subfamily of CYP2B genes is induced by barbiturates (Fujii-Kuriyama et al. Citation1982; Mizukami et al. Citation1983), which are central nervous system depressants, affecting the gamma-aminobutyric acid (GABA) system via modulating GABAA receptors. Hepatic transcription of CYP2B genes is highly inducible by phenobarbital (PB) and PB-like inducers (Adesnik et al. Citation1981; Hardwick et al. Citation1983; Gervot et al. Citation1999) through the constitutive androstane receptor (CAR), pregnane X receptor (PXR) (Sueyoshi et al. Citation1999; Moore et al. Citation2000) and glucocorticoids receptor (GR) (Wang and Negishi Citation2003). There is growing evidence for cross-talk between CAR, PXR, and GR nuclear receptor signaling pathways that regulate cytochrome P450 genes (Kodama et al. Citation2004; Oladimeji et al. Citation2016). CAR and PXR are found in the brain (Lamba, Lamba, Yasuda, et al. Citation2004; Lamba, Yasuda, Lamba, et al. Citation2004; Marini et al. Citation2007; Nannelli et al. Citation2010). Interestingly, active compounds such as phenobarbital, clotrimazole, rifampicin, phenytoin, and 1,4bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) are CYP2B inducers and activators of orphan nuclear receptors CAR and/or PXR (Sueyoshi et al. Citation1999; Moore et al. Citation2000). Phosphorylation of CYP2B leads to posttranslational modification of the enzyme, resulting in its catalytic inactivation and loss of its ability to support monooxygenase activity (Oesch-Bartlomowicz and Oesch Citation2003; ch Citation2005).
The brains of monkeys that have been exposed to ethanol and nicotine at levels comparable to moderate human consumption revealed cellular induction of CYP2B6 and CYP2E1 (Ferguson et al. Citation2013). The pattern of CYP2B6 and CYP2E1 expression detected in the brains of monkeys exposed to ethanol and/or nicotine closely paralleled expression of CYP2B6 and CYP2E1 protein observed in the brains from human alcoholics and smokers (Miksys et al. Citation2002; Howard et al. Citation2003; Miksys et al. Citation2003). Interestingly, ethanol-only exposed monkeys and human alcoholics both presented elevated levels of CYP2B6 protein in the putamen of the striatum and elevated levels of CYP2E1 protein in the dentate gyrus of the hippocampus but unaltered expression of either enzyme in the substantia nigra, i.e. at the beginning of the dopaminergic nigro-striatal pathway, involved in Parkinsonism (Miksys et al. Citation2002; Howard et al. Citation2003; Miksys et al. Citation2003; Ferguson et al. Citation2013), suggesting region-specific regulation. Nicotine exclusively exposed monkeys and human smokers both exhibited elevated levels of CYP2B6 protein in the CA2 region of the hippocampus and elevated levels of CYP2E1 protein in the putamen but had unaltered expression of either enzyme in the caudate of the striatum. Furthermore, increased protein and mRNA levels of CYP2B in rat brain were observed after nicotine pretreatment. These amounts were not affected by inhibition of nicotinic-acetylcholine receptor (nAChR) with an nAChR antagonist, indicating that the mechanism of nicotine induction of brain CYP2B proceeds through increased transcription independently of nAChR activation (Khokhar et al. Citation2010). However, nicotine pretreatment did not alter rat liver CYP2B expression or mRNA levels (Khokhar et al. Citation2010). Some brain-specific sterols, such as dehydroepiandrosterone, induce human CYP2B6 via the constitutive androstane receptor (Kohalmy et al. Citation2007).
Circadian rhythms were shown to control the metabolism of xenobiotics (Preitner et al. Citation2002; Gachon et al. Citation2004, Citation2006; Ripperger Citation2006; Ripperger and Schibler Citation2006). A previous study showed that the clock output genes (i.e. the three PAR basic leucine zipper (bZip) family of transcription factors, Dbp, Hlf (hepatocyte leukemia factor) and Tef (thyrotroph embryonic factor) promote diurnal expression of Cyp2b10 via rhythmic regulation of the CAR receptor, a well-known activator of the enzyme (Gachon et al. Citation2006). Moreover, Claudel et al. (Citation2007) depicted core oscillator manager PARbZip transcription factor, which, when activated, controls expression of CAR and PPARα receptors and induces circadian expression of AhR, contributing to the regulation of circadian drug metabolism.
RNA-editing enzymes (ADAR proteins) and adenosine-to-inosine (A-to-I) editing constitute the dominant type of RNA editing in humans, affecting the majority of human genes (Nishikura Citation2016). RNA editing has essential roles in neurodevelopment and preservation of regular neuronal function (Behm and Öhman Citation2016). Deactivation of ADAR1 or ADAR2 in HepaRG cells reduces expression of CYP2B6 and CYP2C8 mRNA and protein (Nozaki et al. Citation2019). The authors found that decreased expression of the hepatocyte nuclear factor 4α (HNF4α) protein by knockdown of ADARs is correlated with decreased activity of CYPs. mRNA levels of other CYPs, such as CYP2A6, 2C9, 2C19, 2D6, and 2E1, were also decreased by ADAR1 or ADAR2 knockdown, with the exception of CYP3A4 mRNA, which was considerably increased in response to ADAR1 knockdown (Nozaki et al. Citation2019).
CYP2C subfamily
The human CYP2C enzymes together (2C8, 2C9, 2C18, 2C19) metabolize more than half of all commonly administered drugs, arachidonic acid and some steroids. CYP2C9 and CYP2C19 were detected in microsomal fractions of five human brain regions inspected, specifically in the frontal cortex, hippocampus, basal ganglia (the striatum, globus pallidus, and substantia nigra), amygdala (the structure engaged in memory processes, emotional responses, anxiety, aggression), and cerebellum. Notably, CYP2C9 and CYP2C19 together were present mostly within the neuronal soma but with expression extending down the axons and dendrites (Booth Depaz et al. Citation2015). Moreover, CYP2C enzymes in animals have been proposed to contribute to the regulation of basal blood flow in cerebral microcirculation through the generation of epoxyeicosatrienoic acid metabolites from arachidonic acid (Alkayed et al. Citation1996; Iliff et al. Citation2007). Polymorphic CYP2C19, which metabolizes sex hormones and 5-hydroxytryptamine (serotonin), was associated with specific personality traits (such as temperament and character assessment) within healthy Japanese volunteers (Ishii et al. Citation2007). Next, an association between poor metabolizers of CYP2C19 phenotype (CYP2C19*2*2) with a lower level of depressive symptoms was observed in Swedes (Sim et al. Citation2010). Elevated CYP2C19 activity was associated with depression, reduced hippocampal volume, and homeostasis impairment. Studies on a transgenic CYP2C19 mouse model confirmed the involvement of brain CYP2C19 in depression (Jukić et al. Citation2017). Evidence has been reported linking CYP2C9 polymorphism with a reduction in cerebellar volume in epileptic users of phenytoin, a well-established and very effective antiepileptic drug (Twardowschy et al. Citation2013). Another observation suggested that the period of treatment of focal seizures using phenytoin was affected by CYP2C9 gene polymorphism as some patients possess a slow metabolizer variant of CYP2C9 (Veeravigrom et al. Citation2016).
Regulation
The CYP2C cluster resides on chromosome 10q23.33. Four genes are located in sequence order, Tel-CYP2C8-CYP2C9-CYP2C19-CYP2C18-Cen, with a potential opposite arrangement direction versus the centromere and telomere (Gray et al. Citation1995; Walton et al. Citation2005; Pedersen et al. Citation2010; Martis et al. Citation2013). Numerous nuclear receptors have been recognized that mediate xenobiotic-induced transcriptional activation of human CYP2C genes. Constitutive androstane receptor (CAR) and pregnane X receptor (PXR), or both receptors, bind strongly to the identified responsive elements in human CYP2C gene promoters (Jackson et al. Citation2004; Chen et al. Citation2005; Ferguson et al. Citation2005; Jackson et al. Citation2006). The nuclear receptor CAR is responsible for transcriptional activation of CYP2C8, CYP2C9, and CYP2C19 genes (in human primary hepatocytes or HepG2 cells). Notably, CAR was detectable in the human caudate nucleus (Lamba, Yasuda, Lamba, et al. Citation2004), in the hCMEC/D3 cell line, primary cultures of human brain microvessel endothelial cells, and human fetal brain tissue (Chan et al. Citation2011). Other research groups have reported detectable levels of human CAR mRNA in human brain tissue samples (Nishimura et al. Citation2004) and brain glioma cells (Malaplate-Armand et al. Citation2005). Likewise, human PXR has been shown to mediate induction of the CYP2C genes. PXR protein expression has been clearly detected in the human thalamus, pons, and medulla (Nishimura et al. Citation2004; Lamba, Lamba, Yasuda, et al. Citation2004; Miki et al. Citation2005), as well as the hCMEC/D3 cell line, primary cultures of human brain microvessel endothelial cells, and human fetal brain tissue (Chan et al. Citation2011). Cocaine decreases CYP2C8 and CYP2C9 mRNA or protein levels in human U373 MG astrocytoma cells, accompanied by simultaneous downregulation of CAR and nuclear glucocorticoid (GR) receptors (Malaplate-Armand et al. Citation2005). Finally, a comparison of cortex samples from alcoholics and age-matched controls suggested that CYP2C9 expression was increased in individuals with a history of alcohol abuse compared to age- and sex-matched controls (Booth Depaz et al. Citation2015).
RAR-related orphan receptors (RORs) are transcriptional regulators of CYP2C8 in HepG2 cells (Y. Chen et al. Citation2009). Generally, RORα is a nuclear receptor acting as a constitutive activator of transcription (Giguère et al. Citation1994). Interestingly, ROR in the brain seems to be limited to certain neuronal populations of the cerebellum, inferior olive, hippocampus, thalamus, cortex, hypothalamus, and olfactory bulb, as well as in retinal ganglion cells (Ino Citation2004). RORα is highly expressed in Purkinje cells in which it plays a crucial role in differentiation and survival processes (Boukhtouche et al. Citation2006). The nuclear RORs control downstream target genes in a circadian manner, operating to properly gate metabolic events to the appropriate circadian time window (Preitner et al. Citation2002; Sato et al. Citation2004; Akashi and Takumi Citation2005; Guillaumond et al. Citation2005).
An interesting phenomenon of regulation of human CYP2C9 by electrophilic stress was reported by Makia and colleagues (Citation2014). They demonstrated that during electrophoretic mobility shift assays, Fos family proteins (cFos) distinctly interact with the distal activator protein 1 (AP-1) site and Jun family proteins (JunD) with the proximal site. Consequently, the CYP2C9 promoter is stimulated by ectopic expression of cFos and JunD (Makia et al. Citation2014). Neuronal cFos activity is induced in the nuclei of cells under stress (Ceccatelli et al. Citation1989; Chowdhury et al. Citation2000; Kwon et al. Citation2006). Jun family proteins are transcription factors induced after neuronal damage that regulate diverse neuronal injury responses (Herdegen et al. Citation1997; Raivich and Behrens Citation2006). Furthermore, the relaxed chromatin configuration (euchromatin) of neuronal cells facilitates access to DNA, making its structure malleable (Misteli and Soutoglou Citation2009; Casafont et al. Citation2011). These observations suggest that the Makia et al. (Citation2014) hypothesis of DNA looping to activate the CYP2C9 promoter may be used in neurons.
CYP2D subfamily
CYP2D6 exhibits a broad spectrum of substrate specificity proven by in vitro biochemical assays and showing abilities to metabolize 75 drugs, including antidepressants and neuroleptics (Nebert and Russell Citation2002). Brain CYP2D takes part in the local metabolism and analgesic effects of codeine (Zhou et al. Citation2013; McMillan and Tyndale Citation2015, Citation2017), oxycodone (McMillan et al. Citation2019), and tramadol (Wang et al. Citation2015). On the other hand, changes in the activity of brain CYP2D and consequent alterations in metabolism of the antipsychotic drug haloperidol diversely affect acute and chronic side-effects of this drug measured in the rat behavioral models (Miksys et al. Citation2017). The authors suggest that variations in the activity of human brain CYP2D6 may be a risk factor for side-effects of this antipsychotic after acute (e.g. parkinsonism) or chronic (e.g. tardive dyskinesia) treatment. CYP2D6 is one of the most investigated enzymes in the CYP2 family. It is a widely expressed CYP enzyme in brain cells that plays an important role in CNS homeostasis. Shielding neurons against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) toxicity most likely proceeds by CYP2D6 inactivating it to 4-phenyl-1,2,3,6-tetrahydropyridine (Mann and Tyndale Citation2010; Uehara et al. Citation2015; Fernandez-Abascal et al. Citation2018). CYP2D6 catalyzes the synthesis of dopamine from tyramine (Hiroi et al. Citation1998) and of serotonin from 5-methoxytryptamine (Yu et al. Citation2003), and these alternative anabolic pathways of monoaminergic neurotransmitters take place in the brain in vitro (Bromek et al. Citation2011; Haduch et al. Citation2013) and in vivo (Bromek et al. Citation2011; Cheng et al. Citation2013; Haduch et al. Citation2015, Citation2016), suggesting a modulatory role for CYP2D6 in brain function and thereby in essential aspects of brain health and neurotoxicity (Haduch et al. Citation2018, Citation2020). However, the relative contributions of those alternative pathways to the total neurotransmitter synthesis in the brain in vivo at physiological conditions require further investigation. Exposure to drugs and gene polymorphisms of CYP2D6 affect enzyme activity and dopamine formation (Haduch et al. Citation2011; Niwa et al. Citation2017). In brain structures exposed to exogenous melatonin, CYP2D6 supports serotonin synthesis from 5-methoxytryptamine in the brain in vivo, which closes the serotonin-melatonin-serotonin biochemical cycle (Haduch et al. Citation2016).
Studies of cellular localization and regional distribution of CYP2D6 mRNA and protein performed by Siegle et al. (Citation2001) showed that the mRNA was more widely distributed within the brain than the protein. The mRNA was present in both neuronal and glial cells in such structures as the cortex, striatum, globus pallidus, hippocampus, hypothalamus, thalamus, substantia nigra, and cerebellum. In comparison, CYP2D6 protein was primarily localized in large principal neurons, such as pyramidal cells of the cortex and the hippocampus, Purkinje cells of the cerebellum, and in the substantia nigra. A slightly different pattern of CYP2D6 expression in the human brain was observed by Miksys et al. (Citation2002) and Chinta et al. (Citation2002), who also found the enzyme in the striatum. CYP2D6 protein was detected not only in neuronal soma but also in dendrites of Purkinje and cortical neurons. The presence of CYP2D6 protein in the human frontal lobe, hippocampus, and cerebellum was confirmed in the study of Dutheil et al. (Citation2009). Similar to the human brain, CYP2D expression was detected in the abovementioned regions of the monkey brain (Mann et al. Citation2008). In the rodent brain, CYP2D is widely and heterogeneously expressed across brain regions in both neuronal and glial cell populations, including areas unguarded by the BBB, such as the choroid plexus (Norris et al. Citation1996; Miksys et al. Citation2000, Citation2005).
Individuals with poor CYP2D6 metabolism are at a higher risk of Parkinson's disease (Lu et al. Citation2014). In contrast, smokers present higher expression of this enzyme and have a lower risk of neurodegeneration of dopaminergic cells, as in the case of Parkinson’s disease (Mann et al. Citation2008; Yue et al. Citation2008). Furthermore, levels of CYP2D6 seem to be significantly higher in the brains of alcoholics (Miksys et al. Citation2002).
Poor CYP2D6 metabolizers versus extensive metabolizers showed lower vulnerability to psychopathology and greater impulsivity (Peñas-LLedó et al. Citation2009). Increased cerebral activity of CYP2D6 in extensive metabolizers has been noticed in the thalamus and hippocampus, two regions that have high expression of mRNA and CYP2D6 protein (Kirchheiner et al. Citation2011). CYP2D6 is also correlated with neuropsychiatric phenotype differences among patients, indicating its role in anandamide metabolism and implicating this polymorphic enzyme as a potential component of the endocannabinoid system in the brain (Snider et al. Citation2008). In a study concerning the impact of CYP2D6 polymorphism on brain function, brain activation during a working memory task, and an emotional face matching task was measured using fMRI. In both tasks, brain activation increased with increasing CYP2D6 gene activity (Stingl et al. Citation2012). Polymorphic irregularities in CYP2B6 protein levels, expressed in a distinctive zone of the brain within cellular compartments of neurons and astrocytes in humans, reflected specific tissue concentrations and thereby affect the therapeutic and toxic effects of efavirenz in reservoir tissues (Gervot et al. Citation1999; Miksys et al. Citation2003; Michaud et al. Citation2012).
Regulation
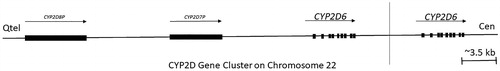
The CYP2D cluster is located on chromosome 22q13.1-2 with only a single functional gene, CYP2D6, as shown in (Koch Citation2004; Ingelman-Sundberg Citation2005). CYP2D6 protein content in the liver is regulated by several transcription factors and fluctuates considerably from person to person, primarily due to its genetic polymorphism. Hepatocyte nuclear factor 4 alpha (HNF4α), a global regulator of genes involved in liver development or liver-specific functions, is the most important hepatic regulatory factor of CYP2D6 (Gonzalez Citation2008). Higher level of HNF4α mRNA expression are significantly correlated with microsomal CYP2D6 enzyme activity (Jover et al. Citation2001; Yang et al. Citation2010).
Figure 5. An outline of the CYP2D gene cluster on human chromosome 22. The cluster contains the CYP2D6 functional gene and two or more highly homologous pseudogenes CYP2D8P and CYP2D7, the gene CYP2D6 is highly polymorphic, and there is substantial variation in allele occurrences between people residing in different geographical location. Arrows and blocks represent the position, relative length, and direction of transcription for each locus, vertical line separate single copy from duplicated type. 5.5% of the Europeans carry more than two active CYP2D6 gene copies and are ultrarapid metabolisers (UMs), whereas the mutation is essentially absent in Asia (adapted from Koch Citation2004; Ingelman-Sundberg Citation2005).
Overexpression of one more regulator CCAAT/enhancer binding protein alpha (C/EBPα) in HepG2 cells increases mRNA levels of CYP2D6, as well as other drug-metabolizing enzymes, such as CYP2B6 and CYP2C9 (Jover et al. Citation1998). Interestingly, C/EBPα is expressed at detectable levels throughout the brain (Protein Atlas version 19.3) and is activated in microglial cells after brain injury. Microglial cells play essential roles in brain injury and repair and are associated with diseases, such as Alzheimer's disease, Creutzfeldt-Jacob disease, multiple sclerosis, the AIDS Dementia Complex, and stroke
Farnesoid X receptor (FXR) activation by sensing bile acids in various tissues or synthetic FXR agonists leads to upregulation of small heterodimer partner (SHP), a transcriptional repressor of CYP2D6 expression (Parks et al. Citation1999; Pan et al. Citation2015). Activation of FXR increases SHP mRNA and protein expression levels in cultured neurons and in brain tissues (Huang et al. Citation2016). Thus, this finding may provide another indication for cerebral regulation of CYP2D6.
A luciferase assay in the SH-SY5Y human neuroblastoma cell line revealed that the PPARγ receptor activated the CYP2D6 gene promoter, while PPARα suppressed its expression (Zhang et al. Citation2018). Furthermore, growth hormone (GH) reduced PPARα mRNA levels and enhanced mRNA levels of CYP2D6 and PPARγ in SH-SY5Y cells. The effect of decreased CYP2D expression was observed in male GHR deficient mice compared to WT mice. Interestingly, brain levels of PPARγ were decreased or unchanged in different areas of the brain in male GHR-/- mice, while brain PPARα expression levels in male GHR-/- mice were significantly greater than those in WT mice brain. Moreover, the results of Zhang et al. (Citation2018) demonstrated that inhibition of cerebellar CYP2D significantly affected spatial learning and exploratory behavior in mice.
There is growing evidence that enhancers are marked in the genome by specific sets of histone modifications, specifically monomethylation of histone 3 lysine 4 (H3K4me) and acetylation of histone 3 lysine 27 (H3K27ac) (Heintzman et al. Citation2007). One study revealed that hypermethylation of CpG islands in the promoter and gene body regions of Cyp2d6 might be crucial for downregulation of this gene in human embryonic stem cell-derived hepatocytes (hESC-Hep). Transcript levels of CYP2D6 were increased after combinatorial inhibition of DNMTs and HDACs (Park et al. Citation2015). DNA methylation is an effective regulator of synaptic function in adult neurons (Feng et al. Citation2010), as well as neuronal activity-induced modification of the neuronal DNA methylome (Guo et al. Citation2011). Expression of cerebral CYP2D enzymes may be regulated by endogenous factors (e.g. hormonal or developmental). Sex steroid hormones affect the expression of brain CYP2D. Testosterone and estrogen positively regulates, while progesterone negatively regulates, CYP2D expression in the brain of female rats (Baum and Strobel Citation1997). Sex hormones can regulate cerebral drug metabolism by governing the expression of an itemized form of microRNAs. Testosterone decreased CYP2D6 activity through upregulation of miRNAs that reduce stability and/or translation of fully or partially sequence-complementary emerging target mRNAs (Landgraf et al. Citation2007; Li et al. Citation2015). Selective regulation of CYP2D6 in SH-SY5Y and U-251, but not in HepG2 cell lines, was also observed. The miR-101 and miR-128-2 bound directly to the 3′-untranslated region of human CYP2D6 mRNA and diminished its functional levels (Li et al. Citation2015). Orchiectomy enhanced brain CYP2D levels in growth hormone receptor (GHR) deficient mice, showing that androgens directly downregulate brain CYP2D expression, while also offering the possibility that brain CYP2D may be affected by growth hormone secretion patterns (Li et al. Citation2015), though pulsatile secreted growth hormone by the pituitary gland does not easily penetrate the BBB (Lai et al. Citation1991; Nyberg and Burman Citation1996).
The potent modulation of CYP2D6 expression by miRNA hsa-miR-370-3p was previously demonstrated (Zeng et al. Citation2017; Zastrozhin et al. Citation2020). miR-370-3p facilitates degradation of CYP2D6 mRNA, decreasing expression and activity of the enzyme. miR-370 is expressed in the brain (Peng et al. Citation2016; Kar et al. Citation2017) and in human neuronal cultures (Peng et al. Citation2016; Evert et al. Citation2018). Recently, an interesting observation was presented by Guo et al. (Citation2020). Overexpression of miR-544 decreases the activity of the epigenetic regulator YY1/PRC2 complex and promotes the transcription and expression of the ten-eleven translocation 2 (TET2) gene, activating critical factors of the serotonergic synapse pathway, CACNA1F and CYP2D6 (Guo et al. Citation2020).
Expression of CYP2D6 in the human brain increases from fetal life to 80 years of age. Three distinct developmental phases of cortical CYP2D6 protein expression were observed; from birth to infancy (0 to 0.9 years old) CYP2D6 expression meaningfully emerges with constant presence, and the second phase with no apparent increase was observed until adulthood (1 to 19 years old). The third phase of a substantial increase in CYP2D6 occurred from 20 to 80 years of age. Moreover, CYP2D6 levels fluctuated among brain regions, with the frontal cortex, caudate, and cerebellum being significantly higher than in the substantia nigra (Mann et al. Citation2012).
Smokers have higher CYP2D6 levels, as assessed in the postmortem brains, while liver CYP2D levels do not differ between smokers and nonsmokers (Miksys et al. Citation2002; Mann et al. Citation2008). Neither rat nor monkey brain CYP2D increases were associated with changes in brain CYP2D mRNA levels, and neither hepatic enzyme activity nor mRNA level was changed, suggesting a brain-specific effect on CYP2D by non-transcriptional regulation (Mann et al. Citation2008; Yue et al. Citation2008; Miller et al. Citation2014). ‘The mechanism of brain CYP2D induction by nicotine is currently unknown but may involve increased protein stability or decreased protein degradation’ (McMillan and Tyndale Citation2018). For instance, nicotine administration dysregulates ubiquitination enzymes in rat brains (Kane et al. Citation2004), and chronic alcohol exposure leads to a significant 25% decreased in the proteasome activity of alcoholic brain tissue (Rezvani et al. Citation2007; Erdozain et al. Citation2014; Pla et al. Citation2014), suggesting one of the possible mechanisms.
Examination of the influence of selected antidepressants and neuroleptics on cytochrome P450 2 D (CYP2D) protein level and activity in the rat brain showed region- and drug-dependent activities, which were changed by prolonged exposure to psychotropics. Chronic thioridazine decreased CYP2D activity and protein level in the nucleus accumbens (i.e. in the ventral striatum) but significantly increased them in the dorsal striatum and cerebellum. Interestingly, neuroleptics decreased the activity with increased CYP2D protein level in the substantia nigra (Haduch et al. Citation2004; Daniel et al. Citation2005). Clozapine also decreased CYP2D activity and protein levels in the nucleus accumbens but significantly enhanced them in the brain stem. In contrast, nefazodone increased enzyme activity, but not protein levels, in the brain stem (Haduch et al. Citation2011, Citation2013). A recent study on the effect of two second-generation antidepressants, escitalopram, and venlafaxine, on the activity of brain and liver CYP2D in a chronic mild stress (CMS) rat model of depression revealed that CMS stimulated CYP2D activity in the hippocampus and evoked the stimulatory effect of antidepressants on enzyme activity in the frontal cortex, hypothalamus and cerebellum (Haduch et al. Citation2018). At the same time, liver CYP2D activity was decreased by the investigated antidepressants. Thus, psychotropics influence CYP2D in the brain, but their effect is different compared to the liver and depends on cerebral cells and network structure. The observed psychotropic–brain CYP2D interactions may affect the metabolism of neurosteroids, monoaminergic neurotransmitters, and biotransformation of drugs, contributing to their pharmacological effects (Haduch et al. Citation2011, Citation2018).
Other CYP2 subfamilies
In the human brain, the most substantial part of expressed metabolizing enzymes is made by CYP2J2, CYP2U1, CYP2E1, and CYP2D6, which represent 90% of total cytochrome P450 enzymes (Dutheil et al. Citation2009). These proteins show varied but selective distribution in different brain regions. For instance, the CYP2U1 protein is present in the frontal lobe at higher levels than in the cerebellum (Karlgren et al. Citation2004; Dutheil et al. Citation2009). The presence of those enzymes was found in both microsomal and mitochondrial fractions and was detected both in neuronal and glial cells in several brain areas (Dutheil et al. Citation2009).
The CYP2E1 gene is placed on chromosome 10 at its longer arm close to the telomere end Ch10q (GeneCards.org). CYP2E1 protein is expressed in pyramidal neurons of the frontal cortex, cerebellum, and hippocampus (Hansson et al. Citation1990; Farin and Omiecinski Citation1993; Upadhya et al. Citation2000; Joshi and Tyndale Citation2006; García‐Suástegui et al. Citation2017). CYP2E1 plays an important role in ethanol oxidation in the brain of rodents (Zimatkin et al. Citation2006). Considering that alcohol dehydrogenase (ADH) is not expressed in the brain, while CYP2E1 is constitutively expressed in various brain regions, it is probable that CYP2E1 is the brain’s primary alcohol metabolizing enzyme (Howard et al. Citation2003).
Dhers et al. (Citation2017) state that cytochrome P450 2U1 is a very specific member of the human P450s family. The CYP2U1 gene is positioned on chromosome 4q25 and spans 18 kb (Dhers et al. Citation2017). This gene shows some structural differences compared to other CYP2 family members, having only five exons and two insertions in the N-terminal region (Karlgren et al. Citation2004; Dhers et al. Citation2017). CYP2U1 metabolizes arachidonic acid, docosahexaenoic acid (DHA), and other long-chain fatty acids. CYP2U1 actively generates highly reactive eicosanoid derivatives, and it is postulated that CYP2U1 plays an essential physiological role in fatty acid signaling processes in both the cerebellum and thymus (Chuang et al. Citation2004). Additionally, expression of CYP2E1 and CYP2U1 proteins in the human prefrontal cortex and amygdala has been confirmed. Furthermore, increased levels of these proteins was identified in the brains of alcoholics and smokers (Toselli et al. Citation2015). Of note, substrate-mediated induction of CYP2E1 and CYP2U1 via protein stabilization in the brain may lead to potent inhibition of other metabolic reactions catalyzed by these enzymes, such as the synthesis and metabolism of neurotransmitters and neuroactive drugs. For example, CYP2U1 metabolizes the tryptophan-derivative indole to oxindole, which is involved in the central nervous system depression associated with liver failure, that is, hepatic encephalopathy (Toselli et al. Citation2015).
The CYP2J2 gene is located on chromosome 1 within the central section of the shorter arm (1p32.1) (GeneCards.org). It has been reported that 2-epoxyeicosatrienoylglycerol (2-AG) is metabolized by CYP2J2 to form 2-11,12-epoxyeicosatrienoic glycerol (EET-G) and 2-14,15-EET-G (McDougle et al. Citation2014). In a rat model, 2-14,15-EET-G was identified in the brain. Both EET-Gs have a high affinity for the cannabinoid receptor (CBR), in particular cannabinoid receptor type 1 (CB1R) (Chen et al. Citation2008), expressed in both peripheral and central nervous systems.
Regulation
Ethanol noticeably increases levels of CYP2E1 protein, mRNA expression, and activity in the hippocampus and cerebellum (Zhong et al. Citation2012). Moreover, generation of ROS induced by ethanol paralleled the additionally elevated CYP2E1 protein levels in the hippocampus, granular layer, and white matter of the cerebellum, as well as the brainstem. The CYP2E1-PPARα axis could be involved in ethanol-induced neurotoxicity by regulating genes related to synaptic function (Na et al. Citation2017).
Parkinson’s disease (PD) is a progressive neurodegenerative disease characterized by loss of dopamine-producing pigmented neurons in the substantia nigra. To detect further PD associated DNA methylation variations, an epigenome-wide association study was performed, and analysis of DNA methylation patterns in putamen samples from postmortem brain tissue of six PD patients was performed. This analysis revealed decreased DNA methylation at the CYP2E1 gene, together with increased expression of the respective CYP2E1 mRNA, suggesting that epigenetically marked cytochrome genes may contribute to PD susceptibility (Kaut et al. Citation2012).
The protein level and activity of CYP2E1 in human liver tissue are negatively correlated with let-7g miRNAs expression (Rieger et al. Citation2013). The let-7 family of miRNAs is one of the most abundant miRNA families in the brain, where it is known to play crucial roles in neurogenesis and neuronal differentiation (Rehfeld et al. Citation2015). Another type of miR-223 reduces the activity of CYP2E1 against the chlorzoxazone 6-hydroxylase substrate in HepG2 cells (Takahashi et al. Citation2014). miR-223 is identified as a regulator of aquaporin-4 (AQP4), a prevalent brain molecule, particularly in astrocyte membranes at the blood-brain and brain-cerebrospinal fluid interfaces. AQP4 is involved in diverse functions, including waste clearance (Nagelhus and Ottersen Citation2013).
Liu et al. (Citation2017) demonstrated the effects of glutamate concentration on the production of epoxyeicosanoids mediated by brain CYP2J (Liu et al. Citation2017). In astrocytes, glutamate increased CYP2J2 mRNA levels in a dose-dependent manner. In contrast, an antagonist of the metabotropic glutamate receptor subtype 5 (mGlu5 receptor) reduced glutamate-induced increases in CYP2J2 mRNA levels. A high dose of glutamate enhanced the binding of CREB to the CYP2J2 promoter. Additionally, inhibition of the MAPK signaling pathway (ERK1/2, p38, and JNK) reduced binding of CREB with the CYP2J2 promoter following glutamate treatment (Liu et al. Citation2017). These results revealed that glutamate in the synaptic cleft might impact the metabolism of endogenous substances in astrocytes via its specific receptor and the MAPK signaling pathway.
Protein levels of CYP2U1, which are principally found in both astrocytes and brain microvessels (Shawahna et al. Citation2011), were induced by glutamate (Yu et al. Citation2019). The authors demonstrated that, similar to CYP1B1, extrasynaptic glutamate from neurotransmitter-mediated signaling pathways in situ affected the metabolism executed via brain CYP2U1 in astrocytes (Yu et al. Citation2019). Downstream genes of CREB and upregulation of CYP2U1 expression by glutamate were observed due to increases in phosphorylated CREB proteins in the nucleus and binding of the CREB protein with the CYP2U1 promoter (Yu et al. Citation2019).
The CYP3 family
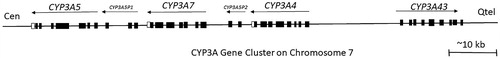
Members of the CYP3 family are responsible for metabolizing 50% of clinical drugs (Evans and Relling Citation1999; Wojnowski Citation2004). The human locus for the CYP3A gene cluster, which includes four genes (CYP3A4, CYP3A5, CYP3A7, and CYP3A43) is located on chromosome 7q21-q22, also containing two pseudogenes (CYP3AP1 and CYP3AP2) () (K. Gellner et al. Citation2001; Chen et al. Citation2009). Each of the four functional genes at the CYP3A locus comprises 13 exons with a processed transcript size of approximately 2 Kb (Klaus Gellner et al. Citation2001). Investigation of CYP3A enzyme activity in the brain is challenging due to the comparatively low content of CYP in the brain and difficulties with its defined localization (Pai et al. Citation2002). Regardless, CYP3A has been identified in a few brain areas at the mRNA, protein, and functional level. The capability of CYP3A enzymes in the brain to metabolize several endogenous and exogenous compounds suggests that they may affect the fate and activity of CNS drugs (Britto and Wedlund Citation1992; Pai et al. Citation2002).
Figure 6. The arrangement of the CYP3A gene cluster with pseudogenes at the CYP3A locus. Abbreviations: Cen - centromere; Qtel - telomere. Exons for genes and pseudogenes are shown as boxes; arrows indicate transcriptional orientation (adapted from Chen et al. Citation2009).
CYP3A subfamily
Currently, the role of the brain CYP3 family is not fully understood. CYP enzyme content in the brain is one-tenth to one-fifteenth of that in the liver (Pai et al. Citation2002). The metabolism of some CYP3A substrates, such as alprazolam by human and rat brain microsomes, to active metabolite α-OH-alprazolam is relatively high. As early as 1993, Farin and Omiecinski used RT-PCR to detect the highest level of CYP3A gene transcripts in the pons, with detectable levels in the cerebellum, frontal lobe, occipital lobe, and red nucleus. Notably, CYP3A mRNA was not detected in the substantia nigra (Farin and Omiecinski Citation1993). Subsequent attempts using better-defined primers showed CYP3A4 and CYP3A5 mRNA expression in the basal ganglia and frontal cortex, while CYP3A5 was also detected in the midbrain (McFadyen et al. Citation1998). Human CYP3A4 is predominantly localized in the Purkinje cell layer in the cerebellum (Pai et al. Citation2002). A brain tumor study showed that antibodies specific for human liver CYP3A4 stained as many as 18% of cells in glioblastomas and 14% of the cells in astrocytomas, whereas less staining was observed in healthy neurons, and staining was not detected in healthy astrocytes (Kirches et al. Citation1999). CYP3A5 is also expressed in the human anterior pituitary gland, specifically in growth hormone-containing cells. The presence of a steroid hydroxylating form of CYP in growth hormone-containing cells may play a role in the comprehensive mechanism to limit steroid hormone feedback regulation of growth hormone secretion (Frohman et al. Citation1992; Murray et al. Citation1995).
Aplrazolam induces expression of the CYP3A43 gene in the human brain cortex, which was significantly higher than in the liver from the same individual when processing drug to its active metabolite (Agarwal et al. Citation2008). CYP3A9 is also highly expressed in the rat brain (Wang et al. Citation1996). These enzymes are found in both neuronal and glial cells in the cell bodies and throughout the cell processes.
In the mouse brain, CYP3A11 and CYP3A13 are found in neuronal populations and, to some extent, in astrocytes. Hagemeyer et al. (Citation2003) demonstrated the inducible ability of these CYP3A isoforms to inactivate testosterone by hydroxylation. The ability of CYP3A enzymes to inactivate testosterone points to their involvement in the regulation of steroid hormone action in the brain. The antiepileptic drug phenytoin induces CYP3a11 in the mouse brain and CYP2B and CYP3A in the rat brain, increasing brain testosterone metabolism in rodents (Rosenbrock et al. Citation1999; Meyer et al. Citation2009). It seems of particular importance that CYP3A expression occurs in nearly all groups of neurons in the hippocampus, which remains widely under the influence of steroid hormones in ontogeny and adulthood. Expression of CYP3A in the brain also suggests that these enzymes might protect the organ against exogenous neurotoxic compounds (Yanagimoto et al. Citation1994; Gokhale et al. Citation1997; Yanagimoto et al. Citation1997; Abu-Qare and Abou-Donia Citation2001; Rosenbrock et al. Citation2001).
Regulation
Many elements and factors have the capacity to regulate the induction of CYP3A. A specific P1 distal promoter of mouse Cyp3a11 transcription is activated by the circadian clock-controlled factor Bmal1, thereby impacting drug detoxification as a function of diurnal time. CYP3A4 is the orthologous gene of Cyp3a11 in humans (Martignoni et al. Citation2006). In vitro analysis provided evidence that human BMAL1 time-dependent regulation of CYP3A4 expression is consistent with the action of the mouse counterpart (Lin et al. Citation2019). Constitutive expression of CYP3A in the liver is synchronized via regular transcription factors, such as HNF1, HNF4, AP1, C/EBPα, C/EBPβ, HNF3γ, and USF1, by bonding to the enhancer module (CLEM4) and the distal enhancer module (XREM) of the CYP3A4 promoter. Xenobiotic-mediated induction of CYP3A is indirect and includes activation of nuclear receptors, such as constitutive androstane receptor (CAR), pregnane X receptor (PXR), vitamin D receptor (VDR), and glucocorticoid receptor (GR). However, PXR is considered the most essential and critical determinant of hepatic CYP3A enzyme activity and expression (Kojima et al. Citation2007; Liu et al. Citation2008). Upon initiation, PXR is translocated to the nucleus, where it heterodimerizes with the retinoid X receptor (RXR) and enhances CYP3A transcription by binding to the AGGTCA-like direct repeat (DR-3) and everted repeat regions (ER-6) on the Cyp3a gene (Liu et al. Citation2008; Taneja et al. Citation2019). In the rat, both CAR and PXR receptors have been reported to be present in low quantities in the cortex and whole brain (Hedlund et al. Citation2001). Unique expression of ligand-activated PXR has been identified in rat brain capillaries regulating a xenobiotic export pump at the blood-brain barrier (Bauer et al. Citation2004). In mouse brain, PXR mediates the inhibitory effect of dexamethasone on the expression of Cyp3a41, while phenobarbital induces expression of Cyp3a11 by activating CAR (Anakk et al. Citation2004). In humans, PXR mRNAs are highly expressed in the thalamus and spinal cord and faintly detectable in the pons and medulla (Lamba, Lamba, Yasuda, et al. Citation2004). Human CYP3A4 is co-expressed in the frontal lobe and thalamus, indicating a poor connection in the expression of CYP3A4 and PXR. CAR was also found to be expressed in the whole brain, however, not in the frontal or temporal lobes, cerebellum, hippocampus, or other specific brain regions (Lamba, Yasuda, Lamba, et al. Citation2004). Robertson et al. (Citation2003) suggested that PXR in the brain may act by shielding it from elevated concentrations of neurosteroids, increasing their metabolism by induction of CYPs and transporters (Robertson et al. Citation2003).
Interestingly, CYP3A9 gene regulation was observed in rat brain and liver controlled by estrogen, exhibiting sexual differences between females and males. Increased concentrations of estrogen enhance CYP3A9 levels (Wang and Strobel Citation1997). Moreover, levels of CYP3A9 mRNA are developmentally regulated, depending on age and sex (Mahnke et al. Citation1997). CYP3A9 mRNA was undetectable by RT-PCR in both male and female livers at the age of 5 weeks or younger, in contrast to the CYP3A9 mRNA, which appeared after the onset of puberty. Two different levels of CYP3A9 mRNA were detected, one for males, which was twice as high in females (Mahnke et al. Citation1997). Sex differences in brain structure and function have long been a subject of consideration, including sex hormone production and development, both prenatally and during adolescence (Kelly et al. Citation1999; Cosgrove et al. Citation2007). Thus, the influence of hormones on cerebral regulation of CYP gene expression is expected.
PXR and GR nuclear receptor expression was shown by immunofluorescence in endothelial cells procured from epileptic brain tissues and in neurons in the human temporal lobe. A more recent study found that mRNA and protein levels of these two receptors were elevated in human brain microvascular endothelial cells of drug-resistant epileptic patients. Interestingly, CYP3A4, CYP2C9, and CYP2E1 were overexpressed in parallel, while CYP2D6 and CYP2C19 were downregulated or absent in endothelial cells (Ghosh et al. Citation2017). The above mechanism might contribute to drug resistance in epilepsy.
Observation of stem cell-derived differentiating neuronal cells from human umbilical cord blood showed that expression of PXR, CAR, AhR, GSTP1-1, and CYPs (CYP1A1, 2B6, 2E1, 3A4) was induced after exposure to the PXR agonist rifampin or organophosphate pesticide monocrotophos (Singh et al. Citation2012). Substantial upregulation of Cyp3a11 and androgen receptor (AR) expression and specific testosterone hydroxylation in neuronal structures of the limbic system (the hippocampus, amygdala, hypothalamus, and thalamus) was observed in mouse brain after phenytoin treatment. Additionally, coherence between CYP3A and AR expression was identified in PC-12 rat derived cells. Thus, induction of Cyp3a11 by phenytoin leads to altered endocrine status within the limbic system, indicating a novel strategy for the improvement of therapy in neurological diseases or brain tumors (Meyer et al. Citation2009).
Summary and discussion
Metabolic processes catalyzed by cytochrome P450 enzymes in the brain can determine the local breakdown of xenobiotics along with the metabolism of endogenous substrates necessary for the proper functioning of the brain and maintaining homeostasis. CYP1, 2, and 3 are primarily responsible for the oxidative metabolism of exogenous substrates and thereby the efficacy of several drugs in brain pharmacotherapy. These enzymes can also be engaged in drug-drug interactions. The metabolism of each active medicinal agent should be determined to assure its bioavailability in specific areas of the brain. A better understanding of the mechanisms of brain CYP enzyme regulation and translation of knowledge to applicable usage may allow for the development of more effective methodologies to treat and prevent CNS diseases. Numerous studies advocate the associations of cerebral CYPs and their endogenous substrates with neurodegenerative diseases, such as Parkinson’s disease (Mann and Tyndale Citation2010; Lu et al. Citation2014; Chattopadhyay et al. Citation2019) or Alzheimer’s disease (Garcia et al. Citation2009; Chace et al. Citation2012; He et al. Citation2012; Jia et al. Citation2016), as well as CYP connection with stress and mental illnesses, such as depression or schizophrenia (Dorado et al. Citation2007; Peñas-LLedó et al. Citation2009; Sim et al. Citation2010; Jukić et al. Citation2017). Cerebral diseases are related to the CYP enzymes that simultaneously metabolize psychotropic drugs used in the therapy of these illnesses and endogenous neuroactive substrates in the brain. Thus, the treatment of cerebral disorders should consider the dual role of some drug-metabolizing enzymes, which may affect not only drug metabolism but also specific endogenous control pathways. Differences in the genome, epigenome, transcriptome, proteome, microRNA, and other molecular components of an individual organism, which can be influenced by various pharmacology of drugs, may lead to inter-individual differences in drug response. Furthermore, knowledge of transporter function in the blood-brain barrier may enhance our capacity to deliver active substances precisely to the brain. Particular attention to the role of CYP enzymes and their metabolic activity and impacting factors (such as the stage of development and genotype exposure to common inducers, including nicotine and ethanol) is likely imperative for the clinical and experimental practice of pharmacology and relative neurotoxicity risk assessment to progress toward personalized treatment of cerebral diseases.
Conclusions
Studies performed hitherto on brain cytochrome P450 indicate that the regulation of CYP gene transcription and CYP protein level and activity is a brain region- and cell-type dependent. Molecular regulation of brain CYP expression and activity differs from that in the liver and is far less known, requiring extensive studies. Nonetheless, levels of some CYPs’ expression/activity in particular brain structures correlate (or coincide) with certain personality traits, the occurrence of neurological diseases, or drug effects. This suggests that the local activity of cytochrome P450 modulates the concentration of endogenous neuroactive substrates and drugs, which may be of epidemiological and therapeutic importance. Therefore, it seems that obtaining an appropriate brain profile of CYP expression/activity (via induction or inhibition), concerning brain structures and endogenous substrates involved in a given illness, may provide an additional mechanism to optimize pharmacotherapy of neurological and psychiatric diseases or to prevent neurodegeneration. This issue needs to be preceded by broadening our knowledge on the biochemical pathways that create the basis for particular brain diseases and on molecular mechanisms that regulate CYP enzyme expression and activity within particular brain areas. Resolving these problems is a challenge for future interdisciplinary research in biochemistry, neuroscience, and neuro- and psychopharmacology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abdelgadir SE, Roselli CE, Choate JV, Resko JA. 1997. Distribution of aromatase cytochrome P450 messenger ribonucleic acid in adult rhesus monkey brains. Biol Reprod. 57(4):772–777.
- Abdel-Razzak Z, Corcos L, Fautrel A, Campion JP, Guillouzo A. 1994. Transforming growth factor-beta 1 down-regulates basal and polycyclic aromatic hydrocarbon-induced cytochromes P-450 1A1 and 1A2 in adult human hepatocytes in primary culture. Mol Pharmacol. 46(6):1100–1110.
- Abu-Qare AW, Abou-Donia MB. 2001. DEET (N,N-diethyl-m-toluamide) alone and in combination with permethrin increased urinary excretion of 6beta-hydroxycortisol in rats, a marker of hepatic CYP3A induction. J Toxicol Environ Health A. 64(5):373–384.
- Acuña-Castroviejo D, Escames G, Venegas C, Díaz-Casado ME, Lima-Cabello E, López LC, Rosales-Corral S, Tan D-X, Reiter RJ. 2014. Extrapineal melatonin: sources, regulation, and potential functions. Cell Mol Life Sci. 71(16):2997–3025.
- Adesnik M, Bar-Nun S, Maschio F, Zunich M, Lippman A, Bard E. 1981. Mechanism of induction of cytochrome P-450 by phenobarbital. J Biol Chem. 256(20):10340–10345.
- Agarwal V, Kommaddi RP, Valli K, Ryder D, Hyde TM, Kleinman JE, Strobel HW, Ravindranath V. 2008. Drug metabolism in human brain: high levels of cytochrome P4503A43 in brain and metabolism of anti-anxiety drug alprazolam to its active metabolite. PLoS One. 3(6):e2337
- Akashi M, Takumi T. 2005. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 12(5):441–448.
- Al Koudsi N, Hoffmann EB, Assadzadeh A, Tyndale RF. 2010. Hepatic CYP2A6 levels and nicotine metabolism: impact of genetic, physiological, environmental, and epigenetic factors. Eur J Clin Pharmacol. 66(3):239–251.
- Alkayed NJ, Birks EK, Hudetz AG, Roman RJ, Henderson L, Harder DR. 1996. Inhibition of brain P-450 arachidonic acid epoxygenase decreases baseline cerebral blood flow. Am J Physiol. 271(4 Pt 2):H1541–1546.
- Anakk S, Kalsotra A, Kikuta Y, Huang W, Zhang J, Staudinger JL, Moore DD, Strobel HW. 2004. CAR/PXR provide directives for Cyp3a41 gene regulation differently from Cyp3a11. Pharmacogenomics J. 4(2):91–101.
- Anandatheerthavarada HK, Williams JF, Wecker L. 1993. The chronic administration of nicotine induces cytochrome P450 in rat brain. J Neurochem. 60(5):1941–1944.
- Annalora AJ, Marcus CB, Iversen PL. 2017. Alternative splicing in the cytochrome P450 superfamily expands protein diversity to augment gene function and redirect human drug metabolism. Drug Metab Dispos. 45(4):375–389.
- Bauer B, Hartz AMS, Fricker G, Miller DS. 2004. Pregnane X receptor up-regulation of P-glycoprotein expression and transport function at the blood-brain barrier. Mol Pharmacol. 66(3):413–419.
- Baulieu E-E, Schumacher M. 2000. Progesterone as a neuroactive neurosteroid, with special reference to the effect of progesterone on myelination. Steroids. 65(10-11):605–612.
- Baum LO, Strobel HW. 1997. Regulation of expression of cytochrome P-450 2D mRNA in rat brain with steroid hormones. Brain Res. 765(1):67–73.
- Bedford FK, Julius D, Ingraham HA. 1998. Neuronal expression of the 5HT 3 serotonin receptor gene requires nuclear factor 1 complexes. J Neurosci. 18(16):6186–6194.
- Behm M, Öhman M. 2016. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 32(3):165–175.
- Bhagwat SV, Boyd MR, Ravindranath V. 2000. Multiple forms of cytochrome P450 and associated monooxygenase activities in human brain mitochondria. Biochem Pharmacol. 59(5):573–582.
- Bhattacharyya MH, DeLuca HF. 1973. The regulation of rat liver calciferol-25-hydroxylase. J Biol Chem. 248(9):2969–2973.
- Bhattacharyya MH, DeLuca HF. 1974. Subcellular location of rat liver calciferol-25-hydroxylase. Arch Biochem Biophys. 160(1):58–62.
- Björkhem I, Lütjohann D, Breuer O, Sakinis A, Wennmalm A. 1997. Importance of a novel oxidative mechanism for elimination of brain cholesterol. Turnover of cholesterol and 24(S)-hydroxycholesterol in rat brain as measured with 18O2 techniques in vivo and in vitro. J Biol Chem. 272(48):30178–30184.
- Booth Depaz IM, Toselli F, Wilce PA, Gillam EMJ. 2015. Differential expression of cytochrome P450 enzymes from the CYP2C subfamily in the human brain. Drug Metab Dispos. 43(3):353–357.
- Boukhtouche F, Janmaat S, Vodjdani G, Gautheron V, Mallet J, Dusart I, Mariani J. 2006. Retinoid-related orphan receptor alpha controls the early steps of Purkinje cell dendritic differentiation. J Neurosci. 26(5):1531–1538.
- Britto MR, Wedlund PJ. 1992. Cytochrome P-450 in the brain. Potential evolutionary and therapeutic relevance of localization of drug-metabolizing enzymes. Drug Metab Dispos. 20(3):446–450.
- Bromek E, Haduch A, Gołembiowska K, Daniel WA. 2011. Cytochrome P450 mediates dopamine formation in the brain in vivo: dopamine formation by brain cytochrome P450 in vivo. J Neurochem. 118(5):806–815.
- Casafont I, Palanca A, Lafarga V, Berciano MT, Lafarga M. 2011. Effect of ionizing radiation in sensory ganglion neurons: organization and dynamics of nuclear compartments of DNA damage/repair and their relationship with transcription and cell cycle. Acta Neuropathol. 122(4):481–493.
- Ceccatelli S, Villar MJ, Goldstein M, Hökfelt T. 1989. Expression of c-Fos immunoreactivity in transmitter-characterized neurons after stress. Proc Natl Acad Sci USA. 86(23):9569–9573.
- Chace C, Pang D, Weng C, Temkin A, Lax S, Silverman W, Zigman W, Ferin M, Lee JH, Tycko B, et al. 2012. Variants in CYP17 and CYP19 Cytochrome P450 genes are associated with onset of Alzheimer's disease in women with down syndrome. J Alzheimers Dis. 28(3):601–612.
- Chacos N, Capdevila J, Falck JR, Manna S, Martin-Wixtrom C, Gill SS, Hammock BD, Estabrook RW. 1983. The reaction of arachidonic acid epoxides (epoxyeicosatrienoic acids) with a cytosolic epoxide hydrolase. Arch Biochem Biophys. 223(2):639–648.
- Chan GNY, Hoque MT, Cummins CL, Bendayan R. 2011. Regulation of P-glycoprotein by orphan nuclear receptors in human brain microvessel endothelial cells. J Neurochem. 118(2):163–175.
- Chattopadhyay M, Chowdhury AR, Feng T, Assenmacher CA, Radaelli E, Guengerich FP, Avadhani NG. 2019. Mitochondrially targeted cytochrome P450 2D6 is involved in monomethylamine-induced neuronal damage in mouse models. J Biol Chem. 294(26):10336–10348.
- Chen H, Howald WN, Juchau MR. 2000. Biosynthesis of all-trans-retinoic acid from all-trans-retinol: Catalysis of all-trans-retinol oxidation by human P-450 cytochromes. Drug Metab Dispos. 28(3):315–322.
- Chen J-K, Chen J, Imig JD, Wei S, Hachey DL, Guthi JS, Falck JR, Capdevila JH, Harris RC. 2008. Identification of Novel Endogenous Cytochrome P450 Arachidonate Metabolites with High Affinity for Cannabinoid Receptors. J Biol Chem. 283(36):24514–24524.
- Chen X, Wang H, Zhou G, Zhang X, Dong X, Zhi L, Jin L, He F. 2009. Molecular population genetics of human CYP3A locus: signatures of positive selection and implications for evolutionary environmental medicine. Environ Health Perspect. 117(10):1541–1548.
- Chen Y, Coulter S, Jetten AM, Goldstein JA. 2009. Identification of human CYP2C8 as a retinoid-related orphan nuclear receptor target gene. J Pharmacol Exp Ther. 329(1):192–201.
- Chen Y, Kissling G, Negishi M, Goldstein JA. 2005. The nuclear receptors constitutive androstane receptor and pregnane x receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther. 314(3):1125–1133.
- Cheng J, Zhen Y, Miksys S, Beyoğlu D, Krausz KW, Tyndale RF, Yu A, Idle JR, Gonzalez FJ. 2013. Potential role of CYP2D6 in the central nervous system. Xenobiotica. 43(11):973–984.
- Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. 2003. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxilase. J Biol Chem. 278(39):38084–38093.
- Cheong RY, Czieselsky K, Porteous R, Herbison AE. 2015. Expression of ESR1 in Glutamatergic and GABAergic Neurons Is Essential for Normal Puberty Onset, Estrogen Feedback, and Fertility in Female Mice. J Neurosci. 35(43):14533–14543.
- Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. 2005. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem Res Toxicol. 18(9):1471–1478.
- Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 460(7254):479–486.
- Chiaro CR, Patel RD, Marcus CB, Perdew GH. 2007. Evidence for an aryl hydrocarbon receptor-mediated cytochrome P450 autoregulatory pathway. Mol Pharmacol. 72(5):1369–1379.
- Chinta SJ, Kommaddi RP, Turman CM, Strobel HW, Ravindranath V. 2005. Constitutive expression and localization of cytochrome P-450 1A1 in rat and human brain: presence of a splice variant form in human brain1. J Neurochem. 93(3):724–736.
- Chinta SJ, Pai HV, Upadhya SC, Boyd MR, Ravindranath V. 2002. Constitutive expression and localization of the major drug metabolizing enzyme, cytochrome P4502D in human brain. Brain Res Mol Brain Res. 103(1–2):49–61.
- Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. 2004. Metabolism of retinoids and arachidonic acid by human and mouse cytochrome P450 1B1. Drug Metab Dispos. 32(8):840–847.
- Chowdhury GM, Fujioka T, Nakamura S. 2000. Induction and adaptation of Fos expression in the rat brain by two types of acute restraint stress. Brain Res Bull. 52(3):171–182.
- Chuang SS, Helvig C, Taimi M, Ramshaw HA, Collop AH, Amad M, White JA, Petkovich M, Jones G, Korczak B. 2004. CYP2U1, a Novel Human Thymus- and Brain-specific Cytochrome P450, Catalyzes omega- and (omega-1)-hydroxylation of fatty acids. J Biol Chem. 279(8):6305–6314.
- Claudel T, Cretenet G, Saumet A, Gachon F. 2007. Crosstalk between xenobiotics metabolism and circadian clock. FEBS Lett. 581(19):3626–3633.
- Cojocaru V, Winn PJ, Wade RC. 2007. The ins and outs of cytochrome P450s. Biochim Biophys Acta BBA - Gen Subj. 1770(3):390–401.
- Cook ND. 2008. The neuron-level phenomena underlying cognition and consciousness: synaptic activity and the action potential. Neuroscience. 153(3):556–570.
- Corchero J, Pimprale S, Kimura S, Gonzalez FJ. 2001. Organization of the CYP1A cluster on human chromosome 15: implications for gene regulation. Pharmacogenetics. 11(1):1–6.
- Cosgrove KP, Mazure CM, Staley JK. 2007. Evolving knowledge of sex differences in brain structure, function, and chemistry. Biol Psychiatry. 62(8):847–855.
- Daly A. CYP2 family [Video file]. In: The Biomedical & Life Sciences Collection, Henry Stewart Talks; 2016 [accessed 2020 Sep 15]. Available from: https://hstalks.com/bs/685/.
- Daniel WA. 2005. The influence of long-term treatment with psychotropic drugs on cytochrome P450: the involvement of different mechanisms. Expert Opin Drug Metab Toxicol. 1(2):203–217.
- Daniel WA, Haduch A, Wójcikowski J. 2005. Inhibition of rat liver CYP2D in vitro and after 1-day and long-term exposure to neuroleptics in vivo-possible involvement of different mechanisms. Eur Neuropsychopharmacol. 15(1):103–110.
- Das ML, Orrenius S, Ernster L. 1968. On the fatty acid and hydrocarbon hydroxylation in rat liver microsomes. Eur J Biochem. 4(4):519–523.
- Dauchy S, Dutheil F, Weaver RJ, Chassoux F, Daumas ‐Duport C, Couraud P-O, Scherrmann J-M, Waziers ID, Declèves X. 2008. ABC transporters, cytochromes P450 and their main transcription factors: expression at the human blood-brain barrier. J Neurochem. 107(6):1518–1528.
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. 2006. The Transcription Factor NFIA Controls the Onset of Gliogenesis in the Developing Spinal Cord. Neuron. 52(6):953–968.
- Dhers L, Ducassou L, Boucher J-L, Mansuy D. 2017. Cytochrome P450 2U1, a very peculiar member of the human P450s family. Cell Mol Life Sci. 74(10):1859–1869.
- Dluzen DF, Lazarus P. 2015. MicroRNA regulation of the major drug-metabolizing enzymes and related transcription factors. Drug Metab Rev. 47(3):320–334.
- Doddapaneni H, Chakraborty R, Yadav JS. 2005. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics. 6:92
- Dorado P, Peñas-Lledó EM, Llerena A. 2007. CYP2D6 polymorphism: implications for antipsychotic drug response, schizophrenia and personality traits. Pharmacogenomics. 8(11):1597–1608.
- Dutheil F, Beaune P, Loriot M-A. 2008. Xenobiotic metabolizing enzymes in the central nervous system: Contribution of cytochrome P450 enzymes in normal and pathological human brain. Biochimie. 90(3):426–436.
- Dutheil F, Dauchy S, Diry M, Sazdovitch V, Cloarec O, Mellottée L, Bièche I, Ingelman-Sundberg M, Flinois J-P, de Waziers I, et al. 2009. Xenobiotic-Metabolizing Enzymes and Transporters in the Normal Human Brain: Regional and Cellular Mapping as a Basis for Putative Roles in Cerebral Function. Drug Metab Dispos. 37(7):1528–1538.
- Erdozain AM, Morentin B, Bedford L, King E, Tooth D, Brewer C, Wayne D, Johnson L, Gerdes HK, Wigmore P, et al. 2014. Alcohol-Related Brain Damage in Humans. PLoS One [Internet]. [accessed 2020 Feb 26] 9(4):e93586.
- Essex MJ, Boyce WT, Hertzman C, Lam LL, Armstrong JM, Neumann SMA, Kobor MS. 2013. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Dev. 84(1):58–75.
- Estabrook RW, Cooper DY, Rosenthal O. 1963. THE light reversible carbon monoxide inhibition of the steroid C21-hydroxylase system of the adrenal cortex. Biochem Z. 338:741–755.
- Evans WE, Relling MV. 1999. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 286(5439):487–491.
- Evert BO, Nalavade R, Jungverdorben J, Matthes F, Weber S, Rajput A, Bonn S, Brüstle O, Peitz M, Krauß S. 2018. Upregulation of miR-370 and miR-543 is associated with reduced expression of heat shock protein 40 in spinocerebellar ataxia type 3. PLoS One. 13(8):e0201794
- Farin FM, Omiecinski CJ. 1993. Regiospecific expression of cytochrome P-450s and microsomal epoxide hydrolase in human brain tissue. J Toxicol Environ Health. 40(2–3):317–335.
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. 2010. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 13(4):423–430.
- Ferguson CS, Miksys S, Palmour RM, Tyndale RF. 2013. Ethanol self-administration and nicotine treatment induce brain levels of CYP2B6 and CYP2E1 in African green monkeys. Neuropharmacology. 72:74–81.
- Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. 2005. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol. 68(3):747–757.
- Fernandez-Abascal J, Ripullone M, Valeri A, Leone C, Valoti M. 2018. β-Naphtoflavone and Ethanol Induce Cytochrome P450 and Protect towards MPP + Toxicity in Human Neuroblastoma SH-SY5Y Cells. IJMS. 19(11):3369.
- Feyereisen R. 2011. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim Biophys Acta BBA - Proteins Proteomics. 1814(1):19–28.
- Finta C, Zaphiropoulos PG. 2000. The human cytochrome P450 3A locus. Gene evolution by capture of downstream exons. Gene. 260(1-2):13–23.
- Frohman LA, Downs TR, Chomczynski P. 1992. Regulation of growth hormone secretion. Front Neuroendocrinol. 13(4):344–405.
- Fujii-Kuriyama Y, Mizukami Y, Kawajiri K, Sogawa K, Muramatsu M. 1982. Primary structure of a cytochrome P-450: coding nucleotide sequence of phenobarbital-inducible cytochrome P-450 cDNA from rat liver. Proc Natl Acad Sci USA. 79(9):2793–2797.
- Fukushima M, Nishii Y, Suzukit M. 1978. Comparative studies on the 25-hydroxylations of cholecalciferol and La-hydroxycholecalciferol in perfused rat liver. Biochem J. 170(3):495–502.
- Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, Schibler U. 2004. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 18(12):1397–1412.
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. 2006. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4(1):25–36.
- Galijatovic A, Beaton D, Nguyen N, Chen S, Bonzo J, Johnson R, Maeda S, Karin M, Guengerich FP, Tukey RH. 2004. The human CYP1A1 gene is regulated in a developmental and tissue-specific fashion in transgenic mice. J Biol Chem. 279(23):23969–23976.
- Garcia ANM, Muniz MTC, Souza e Silva HR, da Silva HA, Athayde-Junior L. 2009. Cyp46 polymorphisms in Alzheimer's disease: a review. J Mol Neurosci. 39(3):342–345.
- Garcia KLP, Coen K, Miksys S, Lê AD, Tyndale RF. 2015. Effect of brain CYP2B inhibition on brain nicotine levels and nicotine self-administration. Neuropsychopharmacol. 40(8):1910–1918.
- García-Suástegui WA, Ramos-Chávez LA, Rubio-Osornio M, Calvillo-Velasco M, Atzin-Méndez JA, Guevara J, Silva-Adaya D. 2017. The role of CYP2E1 in the drug metabolism or bioactivation in the Brain. Romero FJ, editor. Oxid Med Cell Longev. 2017:4680732.
- Gellner K, Eiselt R, Hustert E, Arnold H, Koch I, Haberl M, Deglmann CJ, Burk O, Buntefuss D, Escher S, et al. 2001. Genomic organization of the human CYP3A locus: identification of a new, inducible CYP3A gene. Pharmacogenetics. 11(2):111–121.
- Gervot L, Rochat B, Gautier JC, Bohnenstengel F, Kroemer H, de Berardinis V, Martin H, Beaune P, de Waziers I. 1999. Human CYP2B6: expression, inducibility and catalytic activities. Pharmacogenetics. 9(3):295–306.
- Ghaedi A, Keshavarzi M, Ghafarian Bahraman A, Mohammadi-Bardbori A. 2020. Selective cytochrome P450 1A1 but not 1B1 promoterCpG island DNA methylation by 6-formylindolo[3,2-b]carbazole (FICZ). J Biochem Mol Toxicol. 34(1):e22414.
- Ghosh C, Hossain M, Solanki J, Najm IM, Marchi N, Janigro D. 2017. Overexpression of pregnane X and glucocorticoid receptors and the regulation of cytochrome P450 in human epileptic brain endothelial cells. Epilepsia. 58(4):576–585.
- Giguère V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. 1994. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 8(5):538–553.
- Gilham DE, Cairns W, Paine MJ, Modi S, Poulsom R, Roberts GC, Wolf CR. 1997. Metabolism of MPTP by cytochrome P4502D6 and the demonstration of 2D6 mRNA in human foetal and adult brain by in situ hybridization. Xenobiotica. 27(1):111–125.
- Gokhale MS, Bunton TE, Zurlo J, Yager JD. 1997. Cytochrome P450 isoenzyme activities in cultured rat and mouse liver slices. Xenobiotica. 27(4):341–355.
- Gonzalez FJ. 2008. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet. 23(1):2–7.
- Gonzalez FJ, Kimura S. 2001. Understanding the role of xenobiotic-metabolism in chemical carcinogenesis using gene knockout mice. Mutat Res. 477(1–2):79–87.
- Gotoh O. 1992. Substrate recognition sites in cytochrome P450 Family 2 (CYPB) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 267(1):83–90.
- Gray IC, Nobile C, Muresu R, Ford S, Spurr NK. 1995. A 2.4-megabase physical map spanning the CYP2C gene cluster on chromosome 10q24. Genomics. 28(2):328–332.
- Guillaumond F, Dardente H, Giguère V, Cermakian N. 2005. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 20(5):391–403.
- Guo J, Xiang Q, Xin Y, Huang Y, Zou G, Liu T. 2020. miR-544 promotes maturity and antioxidation of stem cell-derived endothelial like cells by regulating the YY1/TET2 signalling axis. Cell Commun Signal. 18(1):35.
- Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. 2011. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 14(10):1345–1351.
- Haduch A, Bromek E, Daniel WA. 2011. The effect of psychotropic drugs on cytochrome P450 2D (CYP2D) in rat brain. Eur J Pharmacol. 651(1–3):51–58.
- Haduch A, Bromek E, Kot M, Kamińska K, Gołembiowska K, Daniel WA. 2015. The cytochrome P450 2D-mediated formation of serotonin from 5-methoxytryptamine in the brain in vivo: a microdialysis study. J Neurochem. 133(1):83–92.
- Haduch A, Bromek E, Sadakierska-Chudy A, Wójcikowski J, Daniel WA. 2013. The catalytic competence of cytochrome P450 in the synthesis of serotonin from 5-methoxytryptamine in the brain: an in vitro study. Pharmacol Res. 67(1):53–59.
- Haduch A, Bromek E, Wojcikowski J, Go Embiowska K, Daniel WA. 2016. Melatonin supports CYP2D-mediated serotonin synthesis in the brain. Drug Metab Dispos. 44(3):445–452.
- Haduch A, Pukło R, Alenina N, Nikiforuk A, Popik P, Bader M, Daniel WA. 2020. The effect of ageing and cerebral serotonin deficit on the activity of cytochrome P450 2D (CYP2D) in the brain and liver of male rats. Neurochem Int. 141:104884.
- Haduch A, Rysz M, Papp M, Daniel WA. 2018. The activity of brain and liver cytochrome P450 2D (CYP2D) is differently affected by antidepressants in the chronic mild stress (CMS) model of depression in the rat. Biochem Pharmacol. 156:398–405.
- Haduch A, Wójcikowski J, Daniel WA. 2004. Effects of chronic treatment with classic and newer antidepressants and neuroleptics on the activity and level of CYP2D in the rat brain. Pol J Pharmacol. 56(6):857–862.
- Hagemeyer CE, Rosenbrock H, Ditter M, Knoth R, Volk B. 2003. Predominantly neuronal expression of cytochrome P450 isoforms CYP3A11 and CYP3A13 in mouse brain. Neuroscience. 117(3):521–529.
- Hansson T, Tindberg N, Ingelman-Sundberg M, KöHler C. 1990. Regional distribution of ethanol-inducible cytochrome P450 IIE1 in the rat central nervous system. Neuroscience. 34(2):451–463.
- Hardeland R. 2010. Melatonin metabolism in the central nervous system. Curr Neuropharmacol. 8(3):168–181.
- Hardwick JP, Schwalm F, Richardson A. 1983. The effect of phenobarbital on the transcriptional activity of liver. Biochem J. 210(2):599–606.
- He X, Zhang Z, Zhang J-W, Zhou Y, Wu C, Tang M, Hong Z. 2012. An intronic CYP46A1 polymorphism is associated with Alzheimer disease in a Chinese Han population. J Mol Neurosci. 47(3):514–518.
- Hedlund E, Gustafsson J, Warner M. 2001. Cytochrome P450 in the brain: a review. Curr Drug Metab. 2(3):245–263.
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 39(3):311–318.
- Herdegen T, Skene P, Bähr M. 1997. The c-Jun transcription factor-bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 20(5):227–231.
- Hiroi T, Imaoka S, Funae Y. 1998. Dopamine formation from tyramine by CYP2D6. Biochem Biophys Res Commun. 249(3):838–843.
- Hoffman SM, Nelson DR, Keeney DS. 2001. Organization, structure and evolution of the CYP2 gene cluster on human chromosome 19. Pharmacogenetics. 11(8):687–698
- Hojo Y, Hattori T, Enami T, Furukawa A, Suzuki K, Ishii H, Mukai H, Morrison JH, Janssen WGM, Kominami S, et al. 2004. Adult male rat hippocampus synthesizes estradiol from pregnenolone by cytochromes P45017alpha and P450 aromatase localized in neurons. Proc Natl Acad Sci USA. 101(3):865–870.
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. 2012. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22(2):246–258.
- Howard LA, Miksys S, Hoffmann E, Mash D, Tyndale RF. 2003. Brain CYP2E1 is induced by nicotine and ethanol in rat and is higher in smokers and alcoholics. Br J Pharmacol. 138(7):1376–1386.
- Huang C, Wang J, Hu W, Wang C, Lu X, Tong L, Wu F, Zhang W. 2016. Identification of functional farnesoid X receptors in brain neurons. FEBS Lett. 590(18):3233–3242.
- Huang P, Rannug A, Ahlbom E, Håkansson H, Ceccatelli S. 2000. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of cytochrome P450 1A1, the aryl hydrocarbon receptor, and the aryl hydrocarbon receptor nuclear translocator in rat brain and pituitary. Toxicol Appl Pharmacol. 169(2):159–167.
- Iliff JJ, Close LN, Selden NR, Alkayed NJ. 2007. A novel role for P450 eicosanoids in the neurogenic control of cerebral blood flow in the rat. Exp Physiol. 92(4):653–658.
- Ingelman-Sundberg M. 2005. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 5(1):6–13.
- Ino H. 2004. Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J Histochem Cytochem. 52(3):311–323.
- Ishii G, Suzuki A, Oshino S, Shiraishi H, Otani K. 2007. CYP2C19 polymorphism affects personality traits of Japanese females. Neurosci Lett. 411(1):77–80.
- Jackson JP, Ferguson SS, Moore R, Negishi M, Goldstein JA. 2004. The constitutive active/androstane receptor regulates phenytoin induction of Cyp2c29. Mol Pharmacol. 65(6):1397–1404.
- Jackson JP, Ferguson SS, Negishi M, Goldstein JA. 2006. Phenytoin induction of the cyp2c37 gene is mediated by the constitutive androstane receptor. Drug Metab Dispos. 34(12):2003–2010.
- Janmohamed A, Dolphin CT, Phillips IR, Shephard EA. 2001. Quantification and cellular localization of expression in human skin of genes encoding flavin-containing monooxygenases and cytochromes P450. Biochem Pharmacol. 62(6):777–786.
- Jia F, Liu Z, Song N, Du X, Xie J, Jiang H. 2016. The association between CYP46A1 rs4900442 polymorphism and the risk of Alzheimer's disease: A meta-analysis. Neurosci Lett. 620:83–87.
- Jorge-Nebert LF, Jiang Z, Chakraborty R, Watson J, Jin L, McGarvey ST, Deka R, Nebert DW. 2010. Analysis of Human CYP1A1 and CYP1A2 Genes and Their Shared Bidirectional Promoter in Eight World Populations. Hum Mutat. 31(1):27–40.
- Josephy PD, Bibeau KL, Evans DH. 2000. Activation of MeIQ (2-amino-3,4-dimethylimidazo- [4,5-f]quinoline) by sequence variants of recombinant human cytochrome P450 1A2. Environ Mol Mutagen. 35(4):328–335.
- Joshi M, Tyndale RF. 2006. Regional and cellular distribution of CYP2E1 in monkey brain and its induction by chronic nicotine. Neuropharmacology. 50(5):568–575.
- Jover R, Bort R, Gómez-Lechón MJ, Castell JV. 1998. Re-expression of C/EBP alpha induces CYP2B6, CYP2C9 and CYP2D6 genes in HepG2 cells. FEBS Lett. 431(2):227–230.
- Jover R, Bort R, Gómez‐Lechón MJ, Castell JV. 2001. Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: A study using adenovirus-mediated antisense targeting. Hepatology. 33(3):668–675.
- Jukić MM, Opel N, Ström J, Carrillo-Roa T, Miksys S, Novalen M, Renblom A, Sim SC, Peñas-Lledó EM, Courtet P, et al. 2017. Elevated CYP2C19 expression is associated with depressive symptoms and hippocampal homeostasis impairment. Mol Psychiatry. 22(8):1155–1163.
- Kane JK, Konu Ö, Ma JZ, Li MD. 2004. Nicotine coregulates multiple pathways involved in protein modification/degradation in rat brain. Mol Brain Res. 132(2):181–191.
- Kao T-L, Chen Y-L, Kuan Y-P, Chang W-C, Ho Y-C, Yeh S, Jeng L-B, Ma W-L. 2017. Estrogen-Estrogen Receptor α Signaling Facilitates Bilirubin Metabolism in Regenerating Liver Through Regulating Cytochrome P450 2A6 Expression. Cell Transplant. 26(11):1822–1829.
- Kar S, Bali KK, Baisantry A, Geffers R, Samii A, Bertalanffy H. 2017. Genome-Wide Sequencing Reveals MicroRNAs Downregulated in Cerebral Cavernous Malformations. J Mol Neurosci. 61(2):178–188.
- Karlgren M, Backlund M, Johansson I, Oscarson M, Ingelman-Sundberg M. 2004. Characterization and tissue distribution of a novel human cytochrome P450-CYP2U1. Biochem Biophys Res Commun. 315(3):679–685.
- Kaut O, Schmitt I, Wüllner U. 2012. Genome-scale methylation analysis of Parkinson's disease patients' brains reveals DNA hypomethylation and increased mRNA expression of cytochrome P450 2E1. Neurogenetics. 13(1):87–91.
- Kelly SJ, Ostrowski NL, Wilson MA. 1999. Gender differences in brain and behavior: hormonal and neural bases. Pharmacol Biochem Behav. 64(4):655–664.
- Khandwala AS, Kasper CB. 1973. Preferential induction of aryl hydroxylase activity in rat liver nuclear envelope by 3-methylcholanthrene. Biochem Biophys Res Commun. 54(4):1241–1246.
- Khokhar JY, Miksys SL, Tyndale RF. 2010. Rat brain CYP2B induction by nicotine is persistent and does not involve nicotinic acetylcholine receptors. Brain Res. 1348:1–9.
- Khokhar JY, Tyndale RF. 2011. Drug Metabolism within the Brain Changes Drug Response: Selective Manipulation of Brain CYP2B Alters Propofol Effects. Neuropsychopharmacology. 36(3):692–700.
- Kirches E, Scherlach C,V, Bossanyi P, Schneider T, Szibor R, Kutz E, Warich-Kirches M, Dietzmann K. 1999. MGMT- and P450 3A-inhibitors do not sensitize glioblastoma cell cultures against nitrosoureas. Clin Neuropathol. 18(1):1–8.
- Kirchheiner J, Seeringer A, Godoy AL, Ohmle B, Maier C, Beschoner P, Sim E-J, Viviani R. 2011. CYP2D6 in the brain: genotype effects on resting brain perfusion. Mol Psychiatry. 16(3):237:333–341.
- Koch WH. 2004. Technology platforms for pharmacogenomic diagnostic assays. Nat Rev Drug Discov. 3(9):749–761.
- Kodama S, Koike C, Negishi M, Yamamoto Y. 2004. Nuclear Receptors CAR and PXR Cross Talk with FOXO1 To Regulate Genes That Encode Drug-Metabolizing and Gluconeogenic Enzymes. Mol Cell Biol. 24(18):7931–7940.
- Kohalmy K, Tamási V, Kóbori L, Sárváry E, Pascussi J-M, Porrogi P, Rozman D, Prough RA, Meyer UA, Monostory K. 2007. Dehydroepiandrosterone induces human CYP2B6 through the constitutive androstane receptor. Drug Metab Dispos. 35(9):1495–1501.
- Kojima K, Nagata K, Matsubara T, Yamazoe Y. 2007. Broad but Distinct Role of Pregnane X Receptor on the Expression of Individual Cytochrome P450s in Human Hepatocytes. Drug Metab Pharmacokinet. 22(4):276–286.
- Kommaddi RP, Turman CM, Moorthy B, Wang L, Strobel HW, Ravindranath V. 2007. An alternatively spliced cytochrome P4501A1 in human brain fails to bioactivate polycyclic aromatic hydrocarbons to DNA-reactive metabolites. J Neurochem. 102(3):867–877.
- Koskela S, Hakkola J, Hukkanen J, Pelkonen O, Sorri M, Saranen A, Anttila S, Fernandez-Salguero P, Gonzalez F, Raunio H. 1999. Expression of CYP2A genes in human liver and extrahepatic tissues. Biochem Pharmacol. 57(12):1407–1413.
- Kotrbová V, Mrázová B, Moserová M, Martínek V, Hodek P, Hudeček J, Frei E, Stiborová M. 2011. Cytochrome b(5) shifts oxidation of the anticancer drug ellipticine by cytochromes P450 1A1 and 1A2 from its detoxication to activation, thereby modulating its pharmacological efficacy. Biochem Pharmacol. 82(6):669–680.
- Kumar S, Hedges SB. 1998. A molecular timescale for vertebrate evolution. Nature. 392(6679):917–920.
- Kwon M-S, Seo Y-J, Shim E-J, Choi S-S, Lee J-Y, Suh H-W. 2006. The effect of single or repeated restraint stress on several signal molecules in paraventricular nucleus, arcuate nucleus and locus coeruleus. Neuroscience. 142(4):1281–1292.
- Lai ZN, Emtner M, Roos P, Nyberg F. 1991. Characterization of putative growth hormone receptors in human choroid plexus. Brain Res. 546(2):222–226.
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz E. 2004. Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther. 311(2):811–821.
- Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, Schuetz EG. 2004. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 199(3):251–265.
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129(7):1401–1414.
- Lavery DJ, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C. 1999. Circadian expression of the steroid 15 alpha-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 19(10):6488–6499.
- Lee AM, Miksys S, Palmour R, Tyndale RF. 2006. CYP2B6 is expressed in African Green monkey brain and is induced by chronic nicotine treatment. Neuropharmacology. 50(4):441–450.
- Li J, Xie M, Wang X, Ouyang X, Wan Y, Dong G, Yang Z, Yang J, Yue J. 2015. Sex hormones regulate cerebral drug metabolism via brain miRNAs: down-regulation of brain CYP2D by androgens reduces the analgesic effects of tramadol: Regulation of brain CYP2D by androgens via miRNAs. Br J Pharmacol. 172(19):4639–4654.
- Lin Y, Wang S, Zhou Z, Guo L, Yu F, Wu B. 2019. Bmal1 regulates circadian expression of cytochrome P450 3a11 and drug metabolism in mice. Commun Biol. 2(1):1–11.
- Lin YY, Smith LL. 1974. Sterol metabolism XXVIII. Biosynthesis and accumulation of cholest-5-ene-3β, 24-diol (cerebrosterol) in developing rat brain. Biochim Biophys Acta BBA - Lipids Lipid Metab. 348(2):189–196.
- Ling G, Hauer CR, Gronostajski RM, Pentecost BT, Ding X. 2004. Transcriptional regulation of rat CYP2A3 by nuclear factor 1: identification of a novel NFI-A isoform, and evidence for tissue-selective interaction of NFI with the CYP2A3 promoter in vivo. J Biol Chem. 279(27):27888–27895.
- Liu F-J, Song X, Yang D, Deng R, Yan B. 2008. The far and distal enhancers in the CYP3A4 gene co-ordinate the proximal promoter in responding similarly to the pregnane X receptor but differentially to hepatocyte nuclear factor-4alpha. Biochem J. 409(1):243–250.
- Liu M, Zhu Q, Wu J, Yu X, Hu M, Xie X, Yang Z, Yang J, Feng Y-Q, Yue J. 2017. Glutamate affects the production of epoxyeicosanoids within the brain: The up-regulation of brain CYP2J through the MAPK-CREB signaling pathway. Toxicology. 381:31–38.
- Liu Q, Wang W, Zhang Y, Cui Y, Xu S, Li S. 2020. Bisphenol A regulates cytochrome P450 1B1 through miR-27b-3p and induces carp lymphocyte oxidative stress leading to apoptosis. Fish Shellfish Immunol. 102:489–498.
- Lobie PE, García-Aragón J, Lincoln DT, Barnard R, Wilcox JN, Waters MJ. 1993. Localization and ontogeny of growth hormone receptor gene expression in the central nervous system. Brain Res Dev Brain Res. 74(2):225–233.
- Lu AY, Coon MJ. 1968. Role of hemoprotein P-450 in fatty acid omega-hydroxylation in a soluble enzyme system from liver microsomes. J Biol Chem. 243(6):1331–1332.
- Lu J, Shang X, Zhong W, Xu Y, Shi R, Wang X. 2020. New insights of CYP1A in endogenous metabolism: a focus on single nucleotide polymorphisms and diseases. Acta Pharm Sin B. 10(1):91–104.
- Lu Y, Peng Q, Zeng Z, Wang J, Deng Y, Xie L, Mo C, Zeng J, Qin X, Li S. 2014. CYP2D6 phenotypes and Parkinson's disease risk: a meta-analysis. J Neurol Sci. 336(1-2):161–168.
- Lund EG, Guileyardo JM, Russell DW. 1999. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci USA. 96(13):7238–7243.
- Ma X, Idle JR, Krausz KW, Gonzalez FJ. 2005. Metabolism of melatonin by human cytochromes p450. Drug Metab Dispos. 33(4):489–494.
- Mahnke A, Strotkamp D, Roos PH, Hanstein WG, Chabot GG, Nef P. 1997. Expression and inducibility of cytochrome P450 3A9 (CYP3A9) and other members of the CYP3A subfamily in rat liver. Arch Biochem Biophys. 337(1):62–68.
- Makia NL, Surapureddi S, Monostory K, Prough RA, Goldstein JA. 2014. Regulation of Human CYP2C9 Expression by Electrophilic Stress Involves Activator Protein 1 Activation and DNA Looping. Mol Pharmacol. 86(2):125–137.
- Malaplate-Armand C, Ferrari L, Masson C, Visvikis-Siest S, Lambert H, Batt AM. 2005. Down-regulation of astroglial CYP2C, glucocorticoid receptor and constitutive androstane receptor genes in response to cocaine in human U373 MG astrocytoma cells. Toxicol Lett. 159(3):203–211.
- Mann A, Miksys S, Lee A, Mash DC, Tyndale RF. 2008. Induction of the drug metabolizing enzyme CYP2D in monkey brain by chronic nicotine treatment. Neuropharmacology. 55(7):1147–1155.
- Mann A, Miksys SL, Gaedigk A, Kish SJ, Mash DC, Tyndale RF. 2012. The neuroprotective enzyme CYP2D6 increases in the brain with age and is lower in Parkinson's disease patients. Neurobiol Aging. 33(9):2160–2171.
- Mann A, Tyndale RF. 2010. Cytochrome P450 2D6 enzyme neuroprotects against 1-methyl-4-phenylpyridinium toxicity in SH-SY5Y neuronal cells. Eur J Neurosci. 31(7):1185–1193.
- Manzella C, Singhal M, Alrefai WA, Saksena S, Dudeja PK, Gill RK. 2018. Serotonin is an endogenous regulator of intestinal CYP1A1 via AhR. Sci Rep. 8(1):6103.
- Manzella CR, Ackerman M, Singhal M, Ticho AL, Ceh J, Alrefai WA, Saksena S, Dudeja PK, Gill RK. 2020. Serotonin Modulates AhR Activation by Interfering with CYP1A1-Mediated Clearance of AhR Ligands. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 54(1):126–141.
- Marini S, Nannelli A, Sodini D, Dragoni S, Valoti M, Longo V, Gervasi PG. 2007. Expression, microsomal and mitochondrial activities of cytochrome P450 enzymes in brain regions from control and phenobarbital-treated rabbits. Life Sci. 80(10):910–917.
- Martignoni M, Groothuis GMM, de Kanter R. 2006. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2(6):875–894.
- Martis S, Peter I, Hulot J-S, Kornreich R, Desnick RJ, Scott SA. 2013. Multi-ethnic Distribution of Clinically Relevant CYP2C Genotypes and Haplotypes. Pharmacogenomics J. 13(4):369–377.
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. 2009. The epigenetics of sex differences in the brain. J Neurosci. 29(41):12815–12823.
- McDougle DR, Kambalyal A, Meling DD, Das A. 2014. Endocannabinoids Anandamide and 2-Arachidonoylglycerol Are Substrates for Human CYP2J2 Epoxygenase. J Pharmacol Exp Ther. 351(3):616–627.
- McFadyen MCE, Melvin WT, Murray GI. 1998. Regional Distribution of Individual Forms of Cytochrome P450 mRNA in Normal Adult Human Brain. Biochem Pharmacol. 55(6):825–830.
- McMillan DM, Miksys S, Tyndale RF. 2019. Rat brain CYP2D activity alters in vivo central oxycodone metabolism, levels and resulting analgesia. Addict Biol. 24(2):228–238.
- McMillan DM, Tyndale RF. 2015. Nicotine increases codeine analgesia through the induction of brain CYP2D and central activation of codeine to morphine. Neuropsychopharmacol. 40(7):1804–1812.
- McMillan DM, Tyndale RF. 2017. Inducing rat brain CYP2D with nicotine increases the rate of codeine tolerance; predicting the rate of tolerance from acute analgesic response. Biochem Pharmacol. 145:158–168.
- McMillan DM, Tyndale RF. 2018. CYP-mediated drug metabolism in the brain impacts drug response. Pharmacol Ther. 184:189–200.
- Messina ES, Tyndale RF, Sellers EM. 1997. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 282(3):1608–1614.
- Mestres J. 2004. Structure conservation in cytochromes P450. Proteins. 58(3):596–609.
- Meyer RP, Gehlhaus M, Knoth R, Volk B. 2007. Expression and function of cytochrome p450 in brain drug metabolism. Curr Drug Metab. 8(4):297–306.
- Meyer RP, Gehlhaus M, Schwab R, Bürck C, Knoth R, Hagemeyer CE. 2009. Concordant up-regulation of cytochrome P450 Cyp3a11, testosterone oxidation and androgen receptor expression in mouse brain after xenobiotic treatment. J Neurochem. 109(2):670–681.
- Meyer RP, Hagemeyer CE, Knoth R, Kaufmann MR, Volk B. 2006. Anti-epileptic drug phenytoin enhances androgen metabolism and androgen receptor expression in murine hippocampus. J Neurochem. 96(2):460–472.
- Meyer RP, Podvinec M, Meyer UA. 2002. Cytochrome P450 CYP1A1 Accumulates in the Cytosol of Kidney and Brain and Is Activated by Heme. Mol Pharmacol. 62(5):1061–1067.
- Michaud V, Bar-Magen T, Turgeon J, Flockhart D, Desta Z, Wainberg MA. 2012. The Dual Role of Pharmacogenetics in HIV Treatment: Mutations and Polymorphisms Regulating Antiretroviral Drug Resistance and Disposition. Pharmacol Rev. 64(3):803–833.
- Miki Y, Suzuki T, Tazawa C, Blumberg B, Sasano H. 2005. Steroid and xenobiotic receptor (SXR), cytochrome P450 3A4 and multidrug resistance gene 1 in human adult and fetal tissues. Mol Cell Endocrinol. 231(1-2):75–85.
- Miksys S, Hoffmann E, Tyndale RF. 2000. Regional and cellular induction of nicotine-metabolizing CYP2B1 in rat brain by chronic nicotine treatment. Biochem Pharmacol. 59(12):1501–1511.
- Miksys S, Lerman C, Shields PG, Mash DC, Tyndale RF. 2003. Smoking, alcoholism and genetic polymorphisms alter CYP2B6 levels in human brain. Neuropharmacology. 45(1):122–132.
- Miksys S, Rao Y, Hoffmann E, Mash DC, Tyndale RF. 2002. Regional and cellular expression of CYP2D6 in human brain: higher levels in alcoholics: CYP2D6 is higher in brain of human alcoholics. J Neurochem. 82(6):1376–1387.
- Miksys S, Tyndale R. 2013. Cytochrome P450-mediated drug metabolism in the brain. J Psychiatry Neurosci. 38(3):152–163.
- Miksys S, Tyndale RF. 2004. The Unique Regulation of Brain Cytochrome P450 2 (CYP2) family enzymes by drugs and genetics. Drug Metab Rev. 36(2):313–333.
- Miksys S, Wadji FB, Tolledo EC, Remington G, Nobrega JN, Tyndale RF. 2017. Rat brain CYP2D enzymatic metabolism alters acute and chronic haloperidol side-effects by different mechanisms. Prog Neuropsychopharmacol Biol Psychiatry. 78:140–148.
- Miksys SL, Cheung C, Gonzalez FJ, Tyndale RF. 2005. Human CYP2D6 and mouse CYP2DS: organ distribution in a humanized mouse model. Drug Metab Dispos. 33(10):1495–1502.
- Miller RT, Miksys S, Hoffmann E, Tyndale RF. 2014. Ethanol self-administration and nicotine treatment increase brain levels of CYP2D in African green monkeys. Br J Pharmacol. 171(12):3077–3088.
- Misteli T, Soutoglou E. 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 10(4):243–254.
- Mizukami Y, Sogawa K, Suwa Y, Muramatsu M, Fujii-Kuriyama Y. 1983. Gene structure of a phenobarbital-inducible cytochrome P-450 in rat liver. Proc Natl Acad Sci USA. 80(13):3958–3962.
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. 2015. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 86(6):1369–1384.
- Molofsky AV, Hochstim C, Deneen B, Rowitch D. 2013. Mechanisms of Astrocyte Development (Chapter 36). Blood Vessels. 20.
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, et al. 2000. Orphan Nuclear Receptors Constitutive Androstane Receptor and Pregnane X Receptor Share Xenobiotic and Steroid Ligands. J Biol Chem. 275(20):15122–15127.
- Moran JH, Mitchell LA, Bradbury JA, Qu W, Zeldin DC, Schnellmann RG, Grant DF. 2000. Analysis of the cytotoxic properties of linoleic acid metabolites produced by renal and hepatic P450s. Toxicol Appl Pharmacol. 168(3):268–279.
- Murray GI, Pritchard S, Melvin WT, Burke MD. 1995. Cytochrome P450 CYP3A5 in the human anterior pituitary gland. FEBS Lett. 364(1):79–82.
- Muskhelishvili L, Thompson PA, Kusewitt DF, Wang C, Kadlubar FF. 2001. In situ hybridization and immunohistochemical analysis of cytochrome P450 1B1 expression in human normal tissues. J Histochem Cytochem. 49(2):229–236.
- Na S, Li J, Zhang H, Li Y, Yang Z, Zhong Y, Dong G, Yang J, Yue J. 2017. The induction of cytochrome P450 2E1 by ethanol leads to the loss of synaptic proteins via PPARα down-regulation. Toxicology. 385:18–27.
- Nagelhus EA, Ottersen OP. 2013. Physiological roles of aquaporin-4 in brain. Physiol Rev. 93(4):1543–1562.
- Nannelli A, Rossignolo F, Tolando R, Rossato P, Pellegatti M, Longo V, Gervasi PG. 2010. Expression and distribution of CYP3A genes, CYP2B22, and MDR1, MRP1, MRP2, LRP efflux transporters in brain of control and rifampicin-treated pigs. Mol Cell Biochem. 337(1-2):133–143.
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. 2004. Role of Aryl Hydrocarbon Receptor-mediated Induction of the CYP1 Enzymes in Environmental Toxicity and Cancer. J Biol Chem. 279(23):23847–23850.
- Nebert DW, Russell DW. 2002. Clinical importance of the cytochromes P450. The Lancet. 360(9340):1155–1162.
- Nelson DR. 2011. Progress in tracing the evolutionary paths of cytochrome P450. Biochim Biophys Acta. 1814(1):14–18.
- Nishikura K. 2016. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 17(2):83–96.
- Nishimura M, Naito S, Yokoi T. 2004. Tissue-specific mRNA Expression Profiles of Human Nuclear Receptor Subfamilies. Drug Metab Pharmacokinet. 19(2):135–149.
- Niwa T, Shizuku M, Yamano K. 2017. Effect of genetic polymorphism on the inhibition of dopamine formation from p-tyramine catalyzed by brain cytochrome P450 2D6 . Arch Biochem Biophys. 620:23–27.
- Niwa T, Yabusaki Y, Honma K, Matsuo N, Tatsuta K, Ishibashi F, Katagiri M. 1998. Contribution of human hepatic cytochrome P450 isoforms to regioselective hydroxylation of steroid hormones. Xenobiotica. 28(6):539–547.
- Norman BJ, Neal RA. 1976. Examination of the metabolism in vitro of parathion (diethyl p-nitrophenyl phosphorothionate) by rat lung and brain. Biochem Pharmacol. 25(1):37–45.
- Norris PJ, Hardwick JP, Emson PC. 1996. Regional distribution of cytochrome P450 2D1 in the rat central nervous system. J Comp Neurol. 366(2):244–258.
- Nozaki K, Nakano M, Iwakami C, Fukami T, Nakajima M. 2019. RNA Editing Enzymes Modulate the Expression of Hepatic CYP2B6, CYP2C8, and Other Cytochrome P450 Isoforms. Drug Metab Dispos. 47(6):639–647.
- Nyberg F, Burman P. 1996. Growth hormone and its receptors in the central nervous system-location and functional significance. Horm Res. 45(1-2):18–22.
- Obata Y, Castaño Á, Boeing S, Bon-Frauches AC, Fung C, Fallesen T, de Agüero MG, Yilmaz B, Lopes R, Huseynova A, et al. 2020. Neuronal programming by microbiota regulates intestinal physiology. Nature. 578(7794):284–289.
- Oesch-Bartlomowicz B, Oesch F. 2003. Cytochrome-P450 phosphorylation as a functional switch. Arch Biochem Biophys. 409(1):228–234.
- Oesch-Bartlomowicz B, Oesch F. 2005. Phosphorylation of cytochromes P450: first discovery of a posttranslational modification of a drug-metabolizing enzyme. Biochem Biophys Res Commun. 338(1):446–449.
- Ohe T, Hirobe M, Mashino T. 2000. Novel metabolic pathway of estrone and 17beta-estradiol catalyzed by cytochrome P-450. Drug Metab Dispos. 28(2):110–112.
- Oladimeji P, Cui H, Zhang C, Chen T. 2016. Regulation of PXR and CAR by protein-protein interaction and signaling crosstalk. Expert Opin Drug Metab Toxicol. 12(9):997–1010.
- Omura T, Sato R. 1962. A new cytochrome in liver microsomes. J Biol Chem. 237:1375–1376.
- Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. evidence for its hemoprotein nature. J Biol Chem. 239:2370–2378.
- Otyepka M, Skopalík J, Anzenbacherová E, Anzenbacher P. 2007. What common structural features and variations of mammalian P450s are known to date? Biochim Biophys Acta BBA - Gen Subj. 1770(3):376–389.
- Ou‐Yang D-S, Huang S-L, Wang W, Xie H-G, Xu Z-H, Shu Y, Zhou H-H. 2000. Phenotypic polymorphism and gender-related differences of CYP1A2 activity in a Chinese population. Br J Clin Pharmacol. 49(2):145–151.
- Pai HV, Upadhya SC, Chinta SJ, Hegde SN, Ravindranath V. 2002. Differential metabolism of alprazolam by liver and brain cytochrome (P4503A) to pharmacologically active metabolite. Pharmacogenomics J. 2(4):243–258.
- Pan X, Lee Y-K, Jeong H. 2015. Farnesoid X Receptor Agonist Represses Cytochrome P450 2D6 Expression by Upregulating Small Heterodimer Partner. Drug Metab Dispos. 43(7):1002–1007.
- Park H-J, Choi Y-J, Kim JW, Chun H-S, Im I, Yoon S, Han Y-M, Song C-W, Kim H. 2015. Differences in the Epigenetic Regulation of Cytochrome P450 Genes between Human Embryonic Stem Cell-Derived Hepatocytes and Primary Hepatocytes. Aalto-Setala K, editor. PLoS One. 10(7):p. e0132992.
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284(5418):1365–1368.
- Parvez M, Qhanya LB, Mthakathi NT, Kgosiemang IKR, Bamal HD, Pagadala NS, Xie T, Yang H, Chen H, Theron CW, et al. 2016. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci Rep. 6:33099
- Pedersen RS, Brasch-Andersen C, Sim SC, Bergmann TK, Halling J, Petersen MS, Weihe P, Edvardsen H, Kristensen VN, Brøsen K, et al. 2010. Linkage disequilibrium between the CYP2C19*17 allele and wildtype CYP2C8 and CYP2C9 alleles: identification of CYP2C haplotypes in healthy Nordic populations. Eur J Clin Pharmacol. 66(12):1199–1205.
- Penaloza CG, Estevez B, Han DM, Norouzi M, Lockshin RA, Zakeri Z. 2014. Sex-dependent regulation of cytochrome P450 family members Cyp1a1, Cyp2e1, and Cyp7b1 by methylation of DNA. Faseb J. 28(2):966–977.
- Peñas-LLedó EM, Dorado P, Pacheco R, González I, LLerena A. 2009. Relation between CYP2D6 genotype, personality, neurocognition and overall psychopathology in healthy volunteers. Pharmacogenomics. 10(7):1111–1120.
- Peng Z, Wu T, Li Y, Xu Z, Zhang S, Liu B, Chen Q, Tian D. 2016. MicroRNA-370-3p inhibits human glioma cell proliferation and induces cell cycle arrest by directly targeting β-catenin. Brain Res. 1644:53–61.
- Petersen SL, Curran MA, Marconi SA, Carpenter CD, Lubbers LS, McAbee MD. 2000. Distribution of mRNAs encoding the arylhydrocarbon receptor, arylhydrocarbon receptor nuclear translocator, and arylhydrocarbon receptor nuclear translocator-2 in the rat brain and brainstem. J Comp Neurol. 427(3):428–439.
- Phillips JD, Kushner JP, Bergonia HA, Franklin MR. 2011. Uroporphyria in the Cyp1a2-/- mouse. Blood Cells Mol Dis. 47(4):249–254.
- Pla A, Pascual M, Renau-Piqueras J, Guerri C. 2014. TLR4 mediates the impairment of ubiquitin-proteasome and autophagy-lysosome pathways induced by ethanol treatment in brain. Cell Death Dis. 5(2):e1066
- Polimanti R, Jensen KP, Gelernter J. 2017. Phenome-wide association study for CYP2A6 alleles: rs113288603 is associated with hearing loss symptoms in elderly smokers. Sci Rep. 7(1):1–7.
- Poulos TL, Finzel BC, Howard AJ. 1987. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 195(3):687–700.
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 110(2):251–260.
- Raivich G, Behrens A. 2006. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol. 78(6):347–363.
- Raw I, Petragnani N, Nogueira OC. 1960. Studies of electron transport enzymes. IV. Oxidation and reduction of cytochrome b5 by rat liver mitochondria. J Biol Chem. 235:1517–1520.
- Rehfeld F, Rohde AM, Nguyen DTT, Wulczyn FG. 2015. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res. 359(1):145–160.
- Relling MV, Lin JS, Ayers GD, Evans WE. 1992. Racial and gender differences in N-acetyltransferase, xanthine oxidase, and CYP1A2 activities. Clin Pharmacol Ther. 52(6):643–658.
- Ren S, Nguyen L, Wu S, Encinas C, Adams JS, Hewison M. 2005. Alternative splicing of vitamin D-24-hydroxylase: a novel mechanism for the regulation of extrarenal 1,25-dihydroxyvitamin D synthesis. J Biol Chem. 280(21):20604–20611.
- Rezvani K, Teng Y, Shim D, Biasi MD. 2007. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 27(39):10508–10519.
- Rieder CR, Ramsden DB, Williams AC. 1998. Cytochrome P450 1B1 mRNA in the human central nervous system. Mol Pathol. 51(3):138–142.
- Rieger JK, Klein K, Winter S, Zanger UM. 2013. Expression variability of absorption, distribution, metabolism, excretion-related microRNAs in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos. 41(10):1752–1762.
- Ripperger JA. 2006. Mapping of binding regions for the circadian regulators BMAL1 and CLOCK within the mouse Rev-erbalpha gene. Chronobiol Int. 23(1-2):135–142.
- Ripperger JA, Schibler U. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 38(3):369–374.
- Robertson GR, Field J, Goodwin B, Bierach S, Tran M, Lehnert A, Liddle C. 2003. Transgenic Mouse Models of Human CYP3A4 Gene Regulation. Mol Pharmacol. 64(1):42–50.
- Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlic A, Rose PW. 2018. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 34(21):3755–3758.
- Rosenbrock H, Hagemeyer CE, Ditter M, Knoth R, Volk B. 2001. Identification, induction and localization of cytochrome P450s of the 3A-subfamily in mouse brain. Neurotox Res. 3(4):339–349.
- Rosenbrock H, Hagemeyer CE, Singeç I, Knoth R, Volk B. 1999. Testosterone metabolism in rat brain is differentially enhanced by phenytoin-inducible cytochrome P450 isoforms. J Neuroendocrinol. 11(8):597–604.
- Ryan KJ, Engel LL. 1957. Hydroxylation of steroids at carbon 21. J Biol Chem. 225(1):103–114.
- Saeki M, Saito Y, Nagano M, Teshima R, Ozawa S, Sawada J. 2002. mRNA expression of multiple cytochrome p450 isozymes in four types of cultured skin cells. Int Arch Allergy Immunol. 127(4):333–336.
- Sagara Y, Harano T, Omura T. 1978. Characterization of electron transport enzymes in the envelope of rat liver nuclei. J Biochem. 83(3):807–812.
- Sahar S, Sassone-Corsi P. 2012. Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab. 23(1):1–8.
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. 2001. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 15(2):213–225.
- Sasame HA, Ames MM, Nelson SD. 1977. Cytochrome P-450 and NADPH cytochrome c reductase in rat brain: formation of catechols and reactive catechol metabolites. Biochem Biophys Res Commun. 78(3):919–926.
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 43(4):527–537.
- Scandlyn MJ, Stuart EC, Rosengren RJ. 2008. Sex-specific differences in CYP450 isoforms in humans. Expert Opin Drug Metab Toxicol. 4(4):413–424.
- Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, Omenetti S, Henderson CJ, Wolf CR, Nebert DW, et al. 2017. Feedback control of AHR signalling regulates intestinal immunity. Nature. 542(7640):242–245.
- Schilter B, Omiecinski CJ. 1993. Regional distribution and expression modulation of cytochrome P-450 and epoxide hydrolase mRNAs in the rat brain. Mol Pharmacol. 44(5):990–996.
- Schwarz D, Kisselev P, Schunck W-H, Chernogolov A, Boidol W, Cascorbi I, Roots I. 2000. Allelic variants of human cytochrome p450 1A1 (CYP1A1): effect of T461N and I462V substitutions on steroid hydroxylase specificity. Pharmacogenetics. 10(6):519–530.
- Seng JE, Gandy J, Turturro A, Lipman R, Bronson RT, Parkinson A, Johnson W, Hart RW, Leakey JE. 1996. Effects of caloric restriction on expression of testicular cytochrome P450 enzymes associated with the metabolic activation of carcinogens. Arch Biochem Biophys. 335(1):42–52.
- Sezutsu H, Le Goff G, Feyereisen R. 2013. Origins of P450 diversity. Philos Trans R Soc Lond B Biol Sci. 368(1612):20120428.
- Shawahna R, Uchida Y, Declèves X, Ohtsuki S, Yousif S, Dauchy S, Jacob A, Chassoux F, Daumas-Duport C, Couraud P-O, et al. 2011. Transcriptomic and quantitative proteomic analysis of transporters and drug metabolizing enzymes in freshly isolated human brain microvessels. Mol Pharm. 8(4):1332–1341.
- Shehin SE, Stephenson RO, Greenlee WF. 2000. Transcriptional regulation of the human CYP1B1 gene. Evidence for involvement of an aryl hydrocarbon receptor response element in constitutive expression. J Biol Chem. 275(10):6770–6776.
- Shou M, Korzekwa KR, Brooks EN, Krausz KW, Gonzalez FJ, Gelboin HV. 1997. Role of human hepatic cytochrome P450 1A2 and 3A4 in the metabolic activation of estrone. Carcinogenesis. 18(1):207–214.
- Siegle I, Fritz P, Eckhardt K, Zanger UM, Eichelbaum M. 2001. Cellular localization and regional distribution of CYP2D6 mRNA and protein expression in human brain. Pharmacogenetics. 11(3):237–245.
- Sim SC, Nordin L, Andersson TM-L, Virding S, Olsson M, Pedersen NL, Ingelman-Sundberg M. 2010. Association between CYP2C19 polymorphism and depressive symptoms. Am J Med Genet Part B Neuropsychiatr Genet off Publ Int Soc Psychiatr Genet. 153B. (6):1160–1166.
- Sinclair PR, Gorman N, Tsyrlov IB, Fuhr U, Walton HS, Sinclair JF. 1998. Uroporphyrinogen oxidation catalyzed by human cytochromes P450. Drug Metab Dispos. 26(10):1019–1025.
- Singh AK, Kashyap MP, Jahan S, Kumar V, Tripathi VK, Siddiqui MA, Yadav S, Khanna VK, Das V, Jain SK, et al. 2012. Expression and inducibility of cytochrome P450s (CYP1A1, 2B6, 2E1, 3A4) in human cord blood CD34(+) stem cell-derived differentiating neuronal cells. Toxicol Sci off J Soc Toxicol. 129(2):392–410.
- Singh S, Singh K, Patel S, Patel DK, Singh C, Nath C, Singh MP. 2008. Nicotine and caffeine-mediated modulation in the expression of toxicant responsive genes and vesicular monoamine transporter-2 in 1-methyl 4-phenyl-1,2,3,6-tetrahydropyridine-induced Parkinson's disease phenotype in mouse. Brain Res. 1207:193–206.
- Sinues B, Fanlo A, Mayayo E, Carcas C, Vicente J, Arenaz I, Cebollada A. 2008. CYP2A6 activity in a healthy Spanish population: effect of age, sex, smoking, and oral contraceptives. Hum Exp Toxicol. 27(5):367–372.
- Snider NT, Sikora MJ, Sridar C, Feuerstein TJ, Rae JM, Hollenberg PF. 2008. The endocannabinoid anandamide is a substrate for the human polymorphic cytochrome P450 2D6. J Pharmacol Exp Ther. 327(2):538–545.
- Song H-R, Gonzalez-Gomez I, Suh GS, Commins DL, Sposto R, Gilles FH, Deneen B, Erdreich-Epstein A. 2010. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro-Oncol. 12(2):122–132.
- Šrejber M, Navrátilová V, Paloncýová M, Bazgier V, Berka K, Anzenbacher P, Otyepka M. 2018. Membrane-attached mammalian cytochromes P450: An overview of the membrane's effects on structure, drug binding, and interactions with redox partners. J Inorg Biochem. 183:117–136.
- Srinivasan V, Smits M, Spence W, Lowe AD, Kayumov L, Pandi-Perumal SR, Parry B, Cardinali DP. 2006. Melatonin in mood disorders. World J Biol Psychiatry. 7(3):138–151.
- Stahl SM. 2013. Stahl's essential psychopharmacology: neuroscientific basis and practical applications. 4th ed. Cambridge (UK): Cambridge University Press.
- Stapleton G, Steel M, Richardson M, Mason JO, Rose KA, Morris RG, Lathe R. 1995. A Novel Cytochrome P450 Expressed Primarily in Brain. J Biol Chem. 270(50):29739–29745.
- Sterrenburg L, Gaszner B, Boerrigter J, Santbergen L, Bramini M, Elliott E, Chen A, Peeters BWMM, Roubos EW, Kozicz T. 2011. Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One. 6(11):e28128
- Stiborová M, Frei E, Wiessler M, Schmeiser HH. 2001. Human enzymes involved in the metabolic activation of carcinogenic aristolochic acids: evidence for reductive activation by cytochromes P450 1A1 and 1A2. Chem Res Toxicol. 14(8):1128–1137.
- Stingl JC. 2018. Mindful Pharmacogenetics: Drug Dosing for Mental Health. Am J Psychiatry. 175(5):395–397.
- Stingl JC, Esslinger C, Tost H, Bilek E, Kirsch P, Ohmle B, Viviani R, Walter H, Rietschel M, Meyer-Lindenberg A. 2012. Genetic variation in CYP2D6 impacts neural activation during cognitive tasks in humans. Neuroimage. 59(3):2818–2823.
- Su T, Ding X. 2004. Regulation of the cytochrome P450 2A genes. Toxicol Appl Pharmacol. 199(3):285–294.
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. 1999. The Repressed Nuclear Receptor CAR Responds to Phenobarbital in Activating the Human CYP2B6 Gene. J Biol Chem. 274(10):6043–6046.
- Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. 1999. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 216(1):282–296.
- Takahashi JS, Hong H-K, Ko CH, McDearmon EL. 2008. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 9(10):764–775.
- Takahashi K, Oda Y, Toyoda Y, Fukami T, Yokoi T, Nakajima M. 2014. Regulation of cytochrome b5 expression by miR-223 in human liver: effects on cytochrome P450 activities. Pharm Res. 31(3):780–794.
- Taneja G, Maity S, Jiang W, Moorthy B, Coarfa C, Ghose R. 2019. Transcriptomic profiling identifies novel mechanisms of transcriptional regulation of the cytochrome P450 (Cyp) 3a11 gene. Sci Rep. 9(1):1–13.
- Tietz DR, Colthart AM, Pochapsky SS, Pochapsky TC. 2017. Substrate recognition by two different P450s: Evidence for conserved roles in a common fold. Sci Rep. 7(1):1–9.
- Tordjman S, Chokron S, Delorme R, Charrier A, Bellissant E, Jaafari N, Fougerou C. 2017. Melatonin: Pharmacology, Functions and Therapeutic Benefits. Curr Neuropharmacol. 15(3):434–443.
- Toselli F, Booth Depaz IM, Worrall S, Etheridge N, Dodd PR, Wilce PA, Gillam EMJ. 2015. Expression of CYP2E1 and CYP2U1 Proteins in Amygdala and Prefrontal Cortex: Influence of Alcoholism and Smoking. Alcohol Clin Exp Res. 39(5):790–797.
- Toselli F, Dodd PR, Gillam EMJ. 2016. Emerging roles for brain drug-metabolizing cytochrome P450 enzymes in neuropsychiatric conditions and responses to drugs. Drug Metab Rev. 48(3):379–404.
- Trabzuni D, Ramasamy A, Imran S, Walker R, Smith C, Weale ME, Hardy J, Ryten M, North American Brain Expression Consortium 2013. Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun. 4(1):2771.
- Tsuruo Y, Ishimura K, Fujita H, Osawa Y. 1994. Immunocytochemical localization of aromatase-containing neurons in the rat brain during pre- and postnatal development. Cell Tissue Res. 278(1):29–39.
- Turman CM, Hatley JM, Ryder DJ, Ravindranath V, Strobel HW. 2006. Alternative splicing within the human cytochrome P450 superfamily with an emphasis on the brain: The convolution continues. Expert Opin Drug Metab Toxicol. 2(3):399–418.
- Twardowschy CA, Werneck LC, Scola RH, Borgio JG, De Paola L, Silvado C. 2013. The role of CYP2C9 polymorphisms in phenytoin-related cerebellar atrophy. Seizure. 22(3):194–197.
- Tyndale RF, Pianezza ML, Sellers EM. 1999. A common genetic defect in nicotine metabolism decreases risk for dependence and lowers cigarette consumption. Nicotine Tob Res. 1 Suppl 2 Suppl (2):S63–S67. discussion S69-70.
- Ueda R, Iketaki H, Nagata K, Kimura S, Gonzalez FJ, Kusano K, Yoshimura T, Yamazoe Y. 2006. A Common Regulatory Region Functions Bidirectionally in Transcriptional Activation of the Human CYP1A1 and CYP1A2 Genes. Mol Pharmacol. 69(6):1924–1930.
- Uehara S, Uno Y, Inoue T, Murayama N, Shimizu M, Sasaki E, Yamazaki H. 2015. Activation and Deactivation of 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine by Cytochrome P450 Enzymes and Flavin-Containing Monooxygenases in Common Marmosets (Callithrix jacchus). Drug Metab Dispos. 43(5):735–742.
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A. 2015. Tissue-based map of the human proteome. Science. 347(6220):1260419.
- Upadhya SC, Tirumalai PS, Boyd MR, Mori T, Ravindranath V. 2000. Cytochrome P4502E (CYP2E) in brain: constitutive expression, induction by ethanol and localization by fluorescence in situ hybridization. Arch Biochem Biophys. 373(1):23–34.
- Veeravigrom M, Jaroonvanichkul V, Netbaramee W, Phaisarn P, Uyathanarat T. 2016. Phenytoin toxicity in two-month-old Thai infant with CYP2C9 gene polymorphism-A case report. Brain Dev. 38(1):136–138.
- Viitala P, Posti K, Lindfors A, Pelkonen O, Raunio H. 2001. cAMP Mediated Upregulation of CYP2A5 in Mouse Hepatocytes. Biochem Biophys Res Commun. 280(3):761–767.
- Volk B, Hettmannsperger U, Papp T, Amelizad Z, Oesch F, Knoth R. 1991. Mapping of phenytoin-inducible cytochrome P450 immunoreactivity in the mouse central nervous system. Neuroscience. 42(1):215–235.
- Von Bahr C, Ursing C, Yasui N, Tybring G, Bertilsson L, Röjdmark S. 2000. Fluvoxamine but not citalopram increases serum melatonin in healthy subjects - An indication that cytochrome P450 CYP1A2 and CYP2C19 hydroxylate melatonin. Eur J Clin Pharmacol. 56(2):123–127.
- von Richter O, Pitarque M, Rodríguez-Antona C, Testa A, Mantovani R, Oscarson M, Ingelman-Sundberg M. 2004. Polymorphic NF-Y dependent regulation of human nicotine C-oxidase (CYP2A6). Pharmacogenetics. 14(6):369–379.
- Wade RC, Winn PJ, Schlichting I. Sudarko 2004. A survey of active site access channels in cytochromes P450. J Inorg Biochem. 98(7):1175–1182.
- Walsh AA, Szklarz GD, Scott EE. 2013. Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J Biol Chem. 288(18):12932–12943.
- Walton R, Kimber M, Rockett K, Trafford C, Kwiatkowski D, Sirugo G. 2005. Haplotype block structure of the cytochrome P450 CYP2C gene cluster on chromosome 10. Nat Genet. 37(9):915–916.
- Wang H, Donley KM, Keeney DS, Hoffman SMG. 2003. Organization and evolution of the Cyp2 gene cluster on mouse chromosome 7, and comparison with the syntenic human cluster. Environ Health Perspect. 111(15):1835–1842.
- Wang H, Kawashima H, Strobel HW. 1996. cDNA Cloning of a novel CYP3A from rat brain. Biochem Biophys Res Commun. 221(1):157–162.
- Wang H, Negishi M. 2003. Transcriptional Regulation of Cytochrome P450 2B Genes by Nuclear Receptors. Curr Drug Metab. 4(6):515–525.
- Wang H, Strobel HW. 1997. Regulation of CYP3A9 gene expression by estrogen and catalytic studies using cytochrome P450 3A9 expressed in Escherichia coli. Arch Biochem Biophys. 344(2):365–372.
- Wang Q, Han X, Li J, Gao X, Wang Y, Liu M, Dong G, Yue J. 2015. Regulation of cerebral CYP2D alters tramadol metabolism in the brain: interactions of tramadol with propranolol and nicotine. Xenobiotica. 45(4):335–344.
- Warner M, Köhler C, Hansson T, Gustafsson JA. 1988. Regional distribution of cytochrome P-450 in the rat brain: spectral quantitation and contribution of P-450b,e, and P-450c,d. J Neurochem. 50(4):1057–1065.
- White JA, Ramshaw H, Taimi M, Stangle W, Zhang A, Everingham S, Creighton S, Tam S-P, Jones G, Petkovich M. 2000. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc Natl Acad Sci USA. 97(12):6403–6408.
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. 2000. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 5(1):121–131.
- Wojnowski L. 2004. Genetics of the variable expression of CYP3A in humans. Ther Drug Monit. 26(2):192–199.
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'Donnell J, Christensen DJ, Nicholson C, Iliff JJ, et al. 2013. Sleep drives metabolite clearance from the adult brain. Science. 342(6156):373–377.
- Yamazaki H, Shaw PM, Guengerich FP, Shimada T. 1998. Roles of cytochromes P450 1A2 and 3A4 in the oxidation of estradiol and estrone in human liver microsomes. Chem Res Toxicol. 11(6):659–665.
- Yanagimoto T, Itoh S, Sawada M, Hashimoto H, Kamataki T. 1994. Molecular cloning and functional expression of a mouse cytochrome P-450 (Cyp3a-13): examination of Cyp3a-13 enzyme to activate aflatoxin B1 (AFB1). Biochim Biophys Acta. 1201(3):405–410.
- Yanagimoto T, Itoh S, Sawada M, Kamataki T. 1997. Mouse cytochrome P450 (Cyp3a11): predominant expression in liver and capacity to activate aflatoxin B1. Arch Biochem Biophys. 340(2):215–218.
- Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, Gaedigk A, Suver C, Zhong H, Leeder JS, et al. 2010. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 20(8):1020–1036.
- Yasukochi Y, Satta Y. 2015. Molecular Evolution of the CYP2D Subfamily in Primates: Purifying Selection on Substrate Recognition Sites without the Frequent or Long-Tract Gene Conversion. Genome Biol Evol. 7(4):1053–1067.
- Ye W, Chen R, Chen X, Huang B, Lin R, Xie X, Chen J, Jiang J, Deng Y, Wen J. 2019. AhR regulates the expression of human cytochrome P450 1A1 (CYP1A1) by recruiting Sp1. Febs J. 286(21):4215–4231.
- Yoshida R, Nakajima M, Nishimura K, Tokudome S, Kwon J-T, Yokoi T. 2003. Effects of polymorphism in promoter region of human CYP2A6 gene (CYP2A6*9) on expression level of messenger ribonucleic acid and enzymatic activity in vivo and in vitro. Clin Pharmacol Ther. 74(1):69–76.
- Yoshida Y, Noshiro M, Aoyama Y, Kawamoto T, Horiuchi T, Gotoh O. 1997. Structural and evolutionary studies on sterol 14-demethylase P450 (CYP51), the most conserved P450 monooxygenase: II. Evolutionary analysis of protein and gene structures. J Biochem. 122(6):1122–1128.
- Yu A-M, Idle JR, Byrd LG, Krausz KW, Küpfer A, Gonzalez FJ. 2003. Regeneration of serotonin from 5-methoxytryptamine by polymorphic human CYP2D6. Pharmacogenetics. 13(3):173–181.
- Yu X, Wu J, Hu M, Wu J, Zhu Q, Yang Z, Xie X, Feng Y-Q, Yue J. 2019. Glutamate affects the CYP1B1- and CYP2U1-mediated hydroxylation of arachidonic acid metabolism via astrocytic mGlu5 receptor. Int J Biochem Cell Biol. 110:111–121.
- Yue J, Miksys S, Hoffmann E, Tyndale RF. 2008. Chronic nicotine treatment induces rat CYP2D in the brain but not in the liver: an investigation of induction and time course. J Psychiatry Neurosci JPN. 33(1):54–63.
- Yun C-H, Ahn T, Guengerich FP, Yamazaki H, Shimada T. 1999. Phospholipase D Activity of Cytochrome P450 in Human Liver Endoplasmic Reticulum. Arch Biochem Biophys. 367(1):81–88.
- Zastrozhin MS, Smirnov VV, Petukhov AE, Pankratenko EP, Zastrozhina AK, Grishina EA, Ryzhikova KA, Bure IV, Skryabin VY, Vlasovskih RV, et al. 2020. The influence of concentration of micro-RNA hsa-miR-370-3p and CYP2D6*4 on equilibrium concentration of mirtazapine in patients with major depressive disorder. Psychopharmacol Bull. 50(3):58–75.
- Zeng L, Chen Y, Wang Y, Yu L-R, Knox B, Chen J, Shi T, Chen S, Ren Z, Guo L, et al. 2017. MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 140:139–149.
- Zhang F, Li J, Na S, Wu J, Yang Z, Xie X, Wan Y, Li K, Yue J. 2018. The involvement of PPARs in the selective regulation of brain CYP2D by growth hormone. Neuroscience. 379:115–125.
- Zhang J, Ding X. 1998. Identification and characterization of a novel tissue-specific transcriptional activating element in the 5'-flanking region of the CYP2A3 gene predominantly expressed in rat olfactory mucosa . J Biol Chem. 273(36):23454–23462.
- Zhang J, Zhang QY, Guo J, Zhou Y, Ding X. 2000. Identification and functional characterization of a conserved, nuclear factor 1-like element in the proximal promoter region of CYP1A2 gene specifically expressed in the liver and olfactory mucosa. J Biol Chem. 275(12):8895–8902.
- Zhong Y, Dong G, Luo H, Cao J, Wang C, Wu J, Feng Y-Q, Yue J. 2012. Induction of brain CYP2E1 by chronic ethanol treatment and related oxidative stress in hippocampus, cerebellum, and brainstem. Toxicology. 302(2-3):275–284.
- Zhou K, Khokhar JY, Zhao B, Tyndale RF. 2013. First demonstration that brain CYP2D-mediated opiate metabolic activation alters analgesia in vivo. Biochem Pharmacol. 85(12):1848–1855.
- Zhuge J, Yu Y-N. 2004. Three new alternative splicing variants of human cytochrome P450 2D6 mRNA in human extratumoral liver tissue. World J Gastroenterol. 10(22):3356–3360.
- Zimatkin SM, Pronko SP, Vasiliou V, Gonzalez FJ, Deitrich RA. 2006. Enzymatic mechanisms of ethanol oxidation in the brain. Alcohol Clin Exp Res. 30(9):1500–1505.
