?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The regression discontinuity design (RDD) is one of the most credible methods for causal inference that is often regarded as a missing data problem in the potential outcomes framework. However, the methods for missing data such as multiple imputation are rarely used as a method for causal inference. This article proposes multiple imputation regression discontinuity designs (MIRDDs), an alternative way of estimating the local average treatment effect at the cutoff point by multiply-imputing potential outcomes. To assess the performance of the proposed method, Monte Carlo simulations are conducted under 112 different settings, each repeated 5,000 times. The simulation results show that MIRDDs perform well in terms of bias, root mean squared error, coverage, and interval length compared to the standard RDD method. Also, additional simulations exhibit promising results compared to the state-of-the-art RDD methods. Finally, this article proposes to use MIRDDs as a graphical diagnostic tool for RDDs. We illustrate the proposed method with data on the incumbency advantage in U.S. House elections. To implement the proposed method, an easy-to-use software program is also provided.
1. Introduction
Causal inference has been established as a missing data problem using the potential outcomes framework (Rubin Citation1974, p. 689) due to the fundamental problem of causal inference (Holland Citation1986, p. 947). Among the many causal inference techniques for non-experimental data, the regression discontinuity design (RDD) is considered one of the strongest quasi-experimental designs, because RDDs allow us to achieve local randomization around the cutoff point (Lee and Lemieux Citation2015, p. 304; Kim and Steiner Citation2016). The recent literature on RDDs is also based on the potential outcomes framework (Lee Citation2008, p. 678; Imbens and Kalyanaraman Citation2012, p. 934; Calonico, Cattaneo, and Titiunik Citation2014, p. 2298).
In a related vein, Rubin (Citation1987) proposed multiple imputation as a method of dealing with missing data. It is even said, “Multiple imputation is now accepted as the best general method to deal with incomplete data in many fields” (van Buuren Citation2018, p. 30). In both of the literatures of causal inference and missing data analysis, Rubin (Citation1974, Citation1987) played an important role. Indeed, Rubin (Citation2004, pp. 167–168) mentions the possibility of multiply-imputing missing potential outcomes, and recently, some suggestions have been made to use multiple imputation as a method of causal inference (Imbens and Rubin Citation2015, pp. 150–171; van Buuren Citation2018, pp. 241–255). As of today, however, little is known about the performance of multiple imputation to estimate the local average treatment effect (LATE) around the cutoff point.
This article proposes a novel application of multiple imputation for causal inference, which is named multiple imputation regression discontinuity designs (MIRDDs) to be used to estimate the LATE at the cutoff point. Monte Carlo simulations are conducted under 112 different settings, each repeated 5,000 times, and the results of the simulations show that MIRDDs perform well in terms of bias, root mean squared error, coverage, and interval length in comparison with the standard RDD method. Also, additional simulations exhibit promising results compared to the state-of-the-art RDD methods by Calonico, Cattaneo, and Titiunik (Citation2014) and Branson et al. (Citation2019).
Based on the findings that MIRDDs can be used to estimate the LATE at the cutoff, this article further presents a graphical method based on MIRDDs to diagnose the validity of RDD analyses. As Imbens and Lemieux (Citation2008, p. 622) note, graphical analyses are an integral part of RDDs. Therefore, this article will be an important addition to the literature of RDDs in particular and of causal inference in general. We empirically illustrate the proposed method, replicating the renowned application of RDDs to the incumbency advantage in U.S. House elections by Lee (Citation2008).
By way of organization, Sec. 2 reviews the potential outcomes framework that is often used in the causal inference literature, briefly explains how regression discontinuity designs work, and briefly introduces multiple imputation. Section 3 presents MIRDDs, where multiple imputation is used to estimate the LATE at the cutoff point. Section 4 presents Monte Carlo evidence. Section 5 suggests how MIRDDs can be used as a graphical diagnostic tool for RDDs, with an example on the incumbency advantage in U.S. House elections. Section 6 concludes. An easy-to-use software program is also provided in the Appendix.
2. Preliminary concepts in the existing frameworks and methods
This section briefly introduces the framework for causal inference and the mechanism of RDD and MI.
2.1. Review of the potential outcomes framework for causal inference
The modern literature on causal inference has been framed in the Rubin Causal Model, which regards causal inference as a problem of missing data using the concept of potential outcomes (Holland and Rubin Citation1988; Winship and Morgan Citation1999; Little and Rubin Citation2000).
The current study focuses on the causal effect of a binary intervention, which is called a treatment. Let be a treatment for individual
where
means that individuals are not exposed to the treatment while
means that individuals are exposed to the treatment. Also, let
be a continuous outcome variable. The current study focuses on a continuous outcome variable, because the regression discontinuity design is based on a multiple regression model
(Angrist and Pischke Citation2009, p. 253). We will explain
later. Furthermore, let
and
denote the pair of potential outcomes for individual
where
is the outcome without the treatment and
is the outcome with the treatment. Then, the outcome we observe is EquationEq. (1)
(1)
(1) (Imbens and Lemieux Citation2008, p. 616).
(1)
(1)
The individual causal effect (or individual treatment effect) is which is the ultimate interest in causal inference. However, this quantity is unknown, because we only observe
or
for individual
This fact is known to be the “fundamental problem of causal inference” (Holland Citation1986, p. 947). Thus, we usually focus on the average causal effect (or average treatment effect: ATE) as in EquationEq. (2)
(2)
(2) that is “the effect on all individuals (treatment and control)” (Stuart Citation2010, p. 4). Equality in EquationEq. (2)
(2)
(2) follows from the property of expectation that the expectation of a sum is the sum of the expectations.
(2)
(2)
A naïve estimator takes the difference between the mean of observed and the mean of observed
However, it could be confounded by some variables (observed and unobserved).
In the literature of missing data analysis, this naïve estimator is known as available-case analysis (also known as pairwise deletion) that is not a valid method unless data are missing completely at random (MCAR) (van Buuren Citation2018, p. 11; Little and Rubin Citation2020, pp. 61–62).
2.2. Overview of regression discontinuity designs
Let be a continuous variable that determines the assignment of
In the RDD literature, variable
is called the forcing variable (Imbens and Kalyanaraman Citation2012, p. 934; Calonico, Cattaneo, and Titiunik Citation2014, p. 2298), the running variable (Branson et al. Citation2019, p. 15), or simply the assignment variable (Thoemmes, Liao, and Jin Citation2017, p. 341). The current study calls
the forcing variable. In the sharp RDD, which is the focus in the current study, the assignment of a treatment is a deterministic function of
as in EquationEq. (3)
(3)
(3) , where
is a cutoff point.
(3)
(3)
Since deterministically assigns the treatment,
is a confounder. This further means that there is no covariate overlap. Although the propensity score is one of the most often used techniques to address confounding, the propensity score cannot be used in this situation, due to the fact that the overlap assumption is essential for the propensity score (Imbens and Lemieux Citation2008, p. 618; Stuart Citation2010, p. 10; Imbens and Rubin Citation2015, p. 309).
Regression discontinuity designs (RDDs) may be able to address the problem identified above. An intuitive estimator is EquationEq. (4)(4)
(4) , where we take the difference of the expectations between the two groups exactly at the cutoff point. However, there are no observations exactly on the cutoff point.
(4)
(4)
RDDs extend Analysis of Covariance (ANCOVA) as a valid method of causal inference by exploiting the fact that observations just above and below a cutoff can be thought of as a random subsample (Shadish, Cook, and Campbell Citation2002, p. 224). An intuitive explanation is that the difference in around the cutoff is in large part due to random noise. Suppose that
If math test scores for Students A and B are 59 and 60, respectively, then the true math ability of A and B is probably about the same within the margin of error, because no math tests can perfectly measure the true math ability, which further implies math test scores can be considered a function of ability + random noise. Nonetheless, Student A does not receive a treatment while Student B does. Thus, if we focus on a small enough neighborhood around the cutoff, the subsample can be considered a randomly assigned subgroup to the treatment, imitating the randomized experiment. Therefore,
is the only confounding variable in RDDs, which is a remarkable characteristic of RDDs (Angrist and Pischke Citation2009, pp. 257–258). This is why, RDDs are said to be “stronger for causal inference than any design except the randomized experiment” (Shadish, Cook, and Campbell Citation2002, p. 221). Note that we will discuss the issue of bandwidth selection in Sec. 4.3.
The standard model that is used in an RDD is local linear regression, where we “fit linear regression functions to the observations within a distance on either side of the discontinuity point” (Imbens and Lemieux Citation2008, p. 624). This is known to be the local approach as opposed to the global approach such as ANCOVA (Thoemmes, Liao, and Jin Citation2017, p. 344). Indeed, Gelman and Imbens (Citation2019, p. 448) argue that global high-order polynomial regressions should not be used for RDDs; instead, local low-order polynomials such as local linear or local quadratic models are more preferable. Which of the local linear or quadratic models is more appropriate in a given study may be diagnosed by the method presented in Sec. 5 of the current study.
Essentially, RDDs estimate the local average treatment effect (LATE) at the cutoff (Hahn, Todd, and Klaauw Citation2001, p. 204) as in EquationEq. (5)(5)
(5) , which further means that RDDs require the least amount of extrapolation to overcome the lack of covariate overlap (Branson et al. Citation2019, p. 15).
(5)
(5)
Therefore, an RDD has an advantage that it avoids many complications about model specification, such as the selection of variables and functional forms (Hahn, Todd, and Klaauw Citation2001, p. 207).
2.3. Overview of multiple imputation
This section briefly introduces multiple imputation. For a more detailed account on multiple imputation, see Rubin (Citation1987), King et al. (Citation2001), Allison (Citation2002, pp. 27–50), Enders (Citation2010, pp. 164–253), and Carpenter and Kenward (Citation2013).
When some observations are missing in a dataset, it is not possible to conduct statistical analyses. For example, when we have data (1, 3, 4, and NA), the arithmetic mean is (8 + NA)/4, which cannot be numerically computed. Imputation replaces a missing value by some plausible value, so that missing data become pseudo-complete data. For example, our imputed data may look like (1, 3, 4, and 2). Then, the arithmetic mean can be numerically computed as 2.5. However, imputed values are not observed values, but predicted values. This further means that there is uncertainty involved in imputed values. This fact needs to be accounted for in statistical analyses using imputed data.
In theory, multiple imputation replaces a missing value by values (
) independently and randomly drawn from the distribution of missing data. Uncertainty about missing data is reflected in the variation among these
values. For example, our multiply-imputed data may look like (1, 3, 4, and 2), (1, 3, 4, and 0), and (1, 3, 4, and 3). In practice, however, we do not observe missing data by definition, which further means that we do not observe the distribution of missing data.
Under the assumption of missing at random (MAR), where the conditional probability of missing data after controlling for observed data is the same as the probability of observed data (King et al. Citation2001; Allison Citation2002, p. 4; Enders Citation2010, p. 11; Little and Rubin Citation2020, p. 14), multiple imputation constructs the posterior predictive distribution of missing data conditional on observed data. Then, multiple imputation independently and randomly draws simulated values from this posterior predictive distribution. EquationEquation (3)
(3)
(3) states that
is missing when
and
is missing when
Both of these are archetypical MAR. Therefore, it is natural to think that we can apply multiple imputation for the missing portion of
and
In other words, potential outcomes are missing at random in the sense of MAR in the RDD context.
However, it is difficult to randomly draw sufficient statistics from the posterior predictive distribution using the traditional analytical methods. In order to solve this problem, the following three computational algorithms have been proposed in the literature. The traditional algorithm of multiple imputation is the data augmentation (DA) algorithm, which is a Markov chain Monte Carlo (MCMC) technique. For the DA version of multiple imputation, see Schafer (Citation1997, pp. 89–145). An alternative algorithm to DA is the fully conditional specification (FCS) algorithm, which specifies a multivariate distribution by way of a series of conditional densities, through which missing values are imputed given the other variables. Under the compatible conditional densities, FCS is a Gibbs sampler, a type of MCMC (van Buuren Citation2018, p. 120). For the FCS version of multiple imputation, see van Buuren (Citation2018). Another alternative algorithm is the expectation-maximization with bootstrapping (EMB) algorithm, which combines the expectation-maximization (EM) algorithm with the nonparametric bootstrap to create multiply-imputed values of missing data. For the EMB version of multiple imputation, see Honaker and King (Citation2010).
Takahashi (Citation2017) finds that all of the three algorithms are valid methods of multiple imputation for continuous variables, that the EMB algorithm is the fastest among these algorithms, and that the EMB algorithm is most user-friendly. Thus, the analyses in this article are based on the EMB algorithm. Also, Little and Rubin (Citation2020, p. 240) note, “the ML estimates from the bootstrap resamples are asymptotically equivalent to a sample from the posterior distribution of ” Therefore, the EMB version of multiple imputation has the right properties to be used in place of MCMC algorithms (Honaker and King Citation2010, p. 565). The EMB version of multiple imputation is implemented in R-package Amelia (Honaker, King, and Blackwell Citation2011; Honaker, King, and Blackwell Citation2019).
Recently, some authors have contended that multiple imputation can be used as a method of causal inference. For example, Dore et al. (Citation2013, p. S45) suggested a method to impute the missing potential outcomes by combining multiple imputation with the propensity score. Westreich et al. (Citation2015) illustrated the possibility of using multiple imputation as a method of causal inference. Gutman and Rubin (Citation2013, Citation2015) proposed multiple imputation using two splines in subclasses (MITSS) as a procedure to address causal inference from a missing data perspective. Olszewski et al. (Citation2017, p. 108) applied MITSS to immunochemotherapy regimens. Nilsson et al. (Citation2019, p. 825) imputed potential outcomes for causal inference using single imputation. However, little is known about the possibility of using multiple imputation to estimate the LATE at the cutoff. This article fills this gap in the literature.
3. Multiple imputation as a method of estimating the LATE at the cutoff
This section presents multiple imputation regression discontinuity designs (MIRDDS). Following Imbens and Rubin (Citation2015, pp. 150–171) and van Buuren (Citation2018, p. 245), the imputations in EquationEqs. (6)(6)
(6) and Equation(7)
(7)
(7) are alternated to create multiply-imputed values of the missing potential outcomes, where
and
are draws from the parameters of the imputation models, ∼ indicates a random draw from the predictive posterior distribution,
is the unobserved portion of
and
is the observed portion of
(6)
(6)
(7)
(7)
Since multiple imputation alternates EquationEqs. (6)(6)
(6) and Equation(7)
(7)
(7) , the different parameters of the imputation models are estimated for
and
This allows us to adequately estimate the causal impact when the assumption of homogeneity of regression is violated, meaning that
and
have different regression slopes. This is in line with the concept of local regression explained in Sec. 2.
Let be an
dataset, where
is the number of observations and
is the number of variables. In a basic setup, we have two potential outcomes and one forcing variable, i.e.,
and
but we may have additional covariates as well to increase efficiency. Just as in RDDs, the proposed method in the current study also uses dataset
in a small enough neighborhood around the cutoff. In this section, for simplicity, we assume that
is already narrowed down in a small enough neighborhood around the cutoff,
where
is the cutoff point and
is a distance from
For the selection of bandwidth
see Sec. 4.3.
The EMB algorithm works as follows (Honaker and King Citation2010, p. 565). Bootstrap resamples of size are randomly drawn from
with replacement
times (Horowitz Citation2001, pp. 3163–3165; Carsey and Harden Citation2014, p. 215). The variation among these
resamples represents uncertainty about estimation of missing data. Since
is incomplete, the bootstrap resamples are also incomplete. Therefore, the expectation-maximization (EM) algorithm is applied to each of these
bootstrap resamples, which is an iterative method for maximum likelihood estimation (MLE) for incomplete data (Little and Rubin Citation2020, p. 187). In other words, the EM algorithm is used to refine
point estimates of parameter
in the imputation model. Specifically, the mean vector and the variance-covariance matrix are estimated by the EM algorithm as follows. EquationEquation (8)
(8)
(8) is the expectation step that calculates the
-function by averaging the complete-data log-likelihood
over the predictive distribution of missing data, where
is an estimate of the unknown parameter at iteration
EquationEquation (9)
(9)
(9) is the maximization step that finds parameter values at iteration
by maximizing the
-function (Schafer Citation1997, pp. 37–39).
(8)
(8)
(9)
(9)
After obtaining the refined estimates of the mean vector and the variance-covariance matrix, we can calculate
local linear regression imputation models as in EquationEqs. (10)
(10)
(10) and Equation(11)
(11)
(11) , where
in this case. Note that these models can be easily extended to a local quadratic model by including the square term of
in the model. Estimation uncertainty is represented by
regression equations, and fundamental uncertainty by
and
(King et al. Citation2001, p. 54; van Buuren Citation2018, p. 67).
(10)
(10)
(11)
(11)
After multiple imputation, analysis can proceed as if the data are complete, meaning that as if we observe potential outcomes. Let be a point estimate of the LATE based on the
-th multiply-imputed dataset as in EquationEq. (12)
(12)
(12)
(12)
(12)
The combined point estimate of the LATE at the cutoff can be calculated by pooling the results by Rubin’s rule (Rubin Citation1987, p. 76; King et al. Citation2001, p. 53; van Buuren Citation2018, pp. 42–43). The combined point estimate
is EquationEq. (13)
(13)
(13) , which is simply an arithmetic mean of
estimates.
(13)
(13)
The variance of the combined point estimate can be also pooled by Rubin’s rule. Let be
where
is the subsample size in the area
is the average of within-imputation variance as in EquationEq. (14)
(14)
(14) , which is simply an arithmetic mean and represents the usual concept of sampling variability.
is the average of between-imputation variance as in EquationEq. (15)
(15)
(15) , which captures the estimation uncertainty due to missing data.
is the total, combined variance of
as in EquationEq. (16)
(16)
(16) , where
adjusts for the fact that
is finite (Enders Citation2010, pp. 222–223). The 95% confidence interval (
) can be computed as in EquationEq. (17)
(17)
(17) , where
is a t-value with the degrees of freedom
This is just a paired
-test situation; thus, inference is straightforward.
(14)
(14)
(15)
(15)
(16)
(16)
(17)
(17)
4. Monte Carlo simulations
4.1. Settings of population data
We will conduct simulations based on seven different data generation processes to show that MIRDDs can indeed estimate the LATE at the cutoff point. Simulations are conducted in R version 4.0.2. In all of the seven designs, -values are generated by a Beta distribution with parameters
and
based on which the forcing variable is
Also, the error term is
These settings are commonly used in the RDD literature (Imbens and Kalyanaraman Citation2012, p. 950; Calonico, Cattaneo, and Titiunik Citation2014, p. 2314; Branson et al. Citation2019, p20). Simulation runs are repeated 5,000 times for each data generation process just as in Imbens and Kalyanaraman (Citation2012) and Calonico, Cattaneo, and Titiunik (Citation2014). Designs 1 to 4 were first used in the simulations of Imbens and Kalyanaraman (Citation2012), and subsequently used in Calonico, Cattaneo, and Titiunik (Citation2014) and Branson et al. (Citation2019). Designs 5 and 6 were first used in the simulations of Calonico, Cattaneo, and Titiunik (Citation2014), and subsequently used in Branson et al. (Citation2019). Design 7 was first used in Branson et al. (Citation2019).
In design 1, the regression function is a fifth-order polynomial with different coefficients used for and
where
is the cutoff point, as in EquationEq. (18)
(18)
(18) . This setting is based on the study of the incumbency advantage in U.S. House elections by Lee (Citation2008).
(18)
(18)
In design 2, the regression function is quadratic with different coefficients used for and
as in EquationEq. (19)
(19)
(19) .
(19)
(19)
In design 3, the regression function is a fifth-order polynomial with the same coefficients used for and
but the intercept is different by 0.1 points, i.e., a constant average treatment effect, as in EquationEq. (20)
(20)
(20) .
(20)
(20)
In design 4, the regression function is a modification of the third design without as in EquationEq. (21)
(21)
(21) .
(21)
(21)
In design 5, the regression function is a fifth-order polynomial with different coefficients used for and
as in EquationEq. (22)
(22)
(22) . This setting is based on the study on Head Start funding by Ludwig and Miller (Citation2007).
(22)
(22)
In design 6, the regression function is a modification of design 1 with coefficients multiplied by constants in bold-faces to increase curvature as in EquationEq. (23)(23)
(23) .
(23)
(23)
In design 7, the regression function is cubic with different coefficients used for and
as in EquationEq. (24)
(24)
(24) .
(24)
(24)
4.2. Settings of the sample size
While the sample size was set to 500 in Imbens and Kalyanaraman (Citation2012), Calonico, Cattaneo, and Titiunik (Citation2014), and Branson et al. (Citation2019), the sample size in the current study is set to 6,558. This larger sample size is chosen for the following reasons. It is well-known that an RDD generally needs more observations than the standard statistical analyses because using a small neighborhood of the cutoff means that there is not much data (Shadish, Cook, and Campbell Citation2002, p. 212; Angrist and Pischke Citation2009, p. 256). In fact, Lee (Citation2008, p. 691) used the sample size of 6,558, which is the widely cited paper as an exemplar of RDDs. Therefore, in line with the sample size used in Lee (Citation2008), the current study sets the sample size to 6,558, because this is a more realistic sample size for RDDs. Results based on the sample size of 500 are also reported in Sec. 4.8.
4.3. Settings of bandwidths
A bandwidth is calculated by an MSE-optimal bandwidth selector for the difference of regression estimates (mserd), using R-function rdbwselect in R-package rdrobust (Calonico, Cattaneo, and Titiunik Citation2014, p. 2309; Calonico, Cattaneo, and Titiunik Citation2020, p. 4), where MSE stands for the Mean Squared Error. R-package rdrobust is known to be the most advanced RDD package in R, especially for the choice of bandwidth (Thoemmes, Liao, and Jin Citation2017, p. 349). Section 4.7 reports the results based on this bandwidth selector.
We also use other bandwidth estimation methods: An MSE-optimal bandwidth selector for the sum of regression estimates (msesum), a CER-optimal bandwidth selector for the difference of regression estimates (cerrd), and a CER-optimal bandwidth selector for the sum of regression estimates (cersum), where CER stands for the Coverage Error Rate (Calonico, Cattaneo, and Titiunik Citation2020, p. 4). The results for these are reported in the Online Appendix A.
Imbens and Lemieux (Citation2008, p. 633) advise that we should always explore the sensitivity to the choice of bandwidths by using larger or smaller bandwidths than the originally chosen size. Therefore, we also use the bandwidths twice and half of the default size.
4.4. Settings of cutoff points
The cutoff point in Imbens and Kalyanaraman (Citation2012), Calonico, Cattaneo, and Titiunik (Citation2014), and Branson et al. (Citation2019) is all set to However, the performance of RDDs depends on the location of the cutoff point (Shadish, Cook, and Campbell Citation2002, p. 217). Therefore, we also include other cutoff points at
and
where
is the first quartile,
is the median, and
is the third quartile. Section 4.7 reports the results based on
because the results by RDDs are most powerful when the cutoff point is around the center of the distribution of the forcing variable (Shadish, Cook, and Campbell Citation2002, p. 217). Results based on the other cutoff points are reported in the Online Appendix A.
4.5. Evaluation criteria for the simulations
Let be the true population parameter and
be an estimator of
If
in EquationEq. (25)
(25)
(25) , the expected value of
is equal to the true
Then, this estimator
is an unbiased estimator of true parameter
(Mooney Citation1997, p. 59; Gujarati Citation2003, p. 899). Therefore, a good estimator has bias closer to zero.
(25)
(25)
Under some circumstances, however, there is a tradeoff between bias and efficiency of two estimators, because one estimator may have larger bias, but may have smaller variance than another estimator. A slightly biased estimator may be more efficiently scattered around the true parameter than an unbiased estimator. The root mean squared error (RMSE) in EquationEq. (26)(26)
(26) takes a balance between bias and efficiency into account, by measuring the dispersion around the true value of the parameter (Mooney Citation1997, p. 59; Gujarati Citation2003, pp. 901–902; Carsey and Harden Citation2014, pp. 88–89). Therefore, a good estimator has RMSE closer to zero.
(26)
(26)
We are also interested in knowing whether an estimator has a good coverage of the true population parameter
in the sense of the confidence interval (CI), because the data we usually use are a sample, not the population. This can be assessed by computing the empirical coverage probability of the nominal 95% confidence interval, which “is the proportion of simulated samples for which the estimated confidence interval includes the true parameter” (Carsey and Harden Citation2014, p. 93). An estimator with CI closer to the coverage probability of 95% is considered a good estimator.
Also, mean interval length is reported. If the 95% confidence intervals of two estimators both have approximately the empirical coverage probability of 95%, then the shorter interval length is more preferable (van Buuren Citation2018, p. 52).
4.6. Settings of multiple imputation
The DA and FCS versions of multiple imputation are MCMC algorithms that stochastically converge to probability distributions; thus, the assessment of convergence requires expert judgment (Schafer Citation1997, pp. 72, 80; van Buuren Citation2018, pp. 120, 187). On the other hand, the EMB algorithm replaces the complicated process of draws from the posterior density with a bootstrapping algorithm; thus, Markov chains need no burn-in periods (Honaker and King Citation2010, pp. 564–565). Also, the EM algorithm deterministically converges to a point in the parameter space (Schafer Citation1997, p. 80); thus, the assessment of convergence is straightforward. Note that the performance of the EMB algorithm is as good as those of the DA and FCS algorithms, in terms of bias, RMSE, coverage probability, and mean interval length (Takahashi Citation2017).
One thing that needs to be explicitly set in the EMB version of multiple imputation is the number of multiply-imputed datasets While the small number,
was said to be enough in the past (Rubin Citation1987, p. 114; Schafer Citation1997, p. 107), the more recent literature recommends to use a larger number,
(Graham, Olchowski, and Gilreath Citation2007; Bodner Citation2008; van Buuren Citation2018, p. 58). Multiple imputation is a simulation-based technique. Therefore, in theory, the larger
is, the better-off we are. Considering the computational burden in Monte Carlo simulation from a practical point of view as well, this study sets
In the software provided in the Appendix,
is set to 100 by default, but the user can freely change this number.
Other settings use the defaults of R-package Amelia, but in a particular application, these settings may be changed. For this issue, we may assess the sensitivity of the multiple imputation settings by using the methods introduced in Sec. 5 and the software introduced in the Appendix.
4.7. Simulation results
presents the results of the simulations at the the median of the forcing variable. Full results using the other cutoff points and the other bandwidth selection techniques are reported in the Online Appendix A.
Table 1. (a). Results of the simulation for Populations 1 to 4 at the
Table 1. (b). Results of the simulation for Populations 5 to 7 at the
In , Bias, RMSE, Coverage, CI Length, and Sub-Sample Size are reported. Bias and RMSE are defined in EquationEqs. (25)(25)
(25) and Equation(26)
(26)
(26) , respectively. Coverage is the empirical coverage probability of the nominal 95% confidence interval. CI Length is mean interval length of the 95% confidence interval. Sub-Sample Size is the average number of observations used to estimate the LATE at the cutoff point.
In , Comp is the ideal but unavailable situation, where we have both potential outcomes and
meaning that the data are complete. Since Comp does not need to estimate a model, the bandwidth can be as narrow as possible. In this simulation, Comp uses one-tenth of the bandwidth automatically selected by R-function rdbwselect. Naïve is the naïve estimator that simply takes a difference between the mean of observed
and the mean of observed
MIRDD is the proposed method based on multiple imputation regression discontinuity designs explained in Sec. 3. RDD is the regression discontinuity design.
In all of the situations, Comp is unbiased and most efficient with the coverage probability being approximately 95%, and Naïve is biased and least efficient with the coverage probability being lower than 95%. Essentially, these are the upper and lower bounds of performance. The overall conclusion is that MIRDD (the proposed method) performs as equally well as RDD. The results generally hold true in the other situations as well (See the Online Appendix A).
summarizes the overall results of the simulations. Reported values are the percentage that each method outperforms the other method. Overall, MIRDD is slightly more biased, but it is more balanced in terms of bias and efficiency than RDD. Depending on the cutoff points and the population types, MIRDD sometimes outperforms RDD, while RDD sometimes outperforms MIRDD. In other words, MIRDD and RDD have different strengths and weaknesses; thus, they may complement each other. Note, however, that the differences in bias and RMSE between MIRDD and RDD are generally very small. For the detailed results, see the Online Appendix A.
Table 2. Overall results. The percentage each method wins against the other method. Larger numbers indicate better results.
One thing that is consistently true in all of the simulations is that a narrower bandwidth leads to a better performance outcome. This finding is consistent with Calonico, Cattaneo, and Titiunik (Citation2014, p. 2301), who argue that the MSE-optimal bandwidth tends to be too large. This indicates that, even though this bandwidth is said to be optimal, this is still an optimal bandwidth as a starting point. Thus, in real applications, we should diagnose whether the automatically selected bandwidth is adequate or not.
4.8. Comparison of MIRDDs and the state-of-the-art RDDs
This section replicates the simulations conducted by Branson et al. (Citation2019, p. 27) to compare the performance of MIRDDs against those of the state-of-the-art methods of RDDs by Calonico, Cattaneo, and Titiunik (Citation2014) and Branson et al. (Citation2019). This comparison is possible because the seven populations used in the current study are also used in Branson et al. (Citation2019) with the same criteria of bias, RMSE, coverage, and mean interval length.
In , LLR is local linear regression (standard RDD), Robust LLR is robust local linear regression proposed by Calonico, Cattaneo, and Titiunik (Citation2014) to adjust for the bias in RDDs, GPR is Gaussian process regression proposed by Branson et al. (Citation2019) to make the fit of RDD flexible, RDD is the same RDD method used in of the current study, and MIRDD is the proposed method in the current study. The results for LLR, Robust LLR, and GPR are directly taken from in Branson et al. (Citation2019, p. 27). Essentially, LLR and RDD are the same method; therefore, the differences we observe between them are due to simulation error.
Table 3. Results of the replication of the simulations in Branson et al. (Citation2019).
In all of the situations, robust LLR and MIRDD are competitive, meaning that their strengths and weaknesses depend on population types. On the other hand, GPR is clearly inferior to MIRDD. Readers may be slightly concerned about the performance of MIRDD in population 2; however, if we use the half-size of the chosen bandwidth in , the performance of MIRDD is satisfactory. In a real application, this decision can be further assessed through the diagnostic method introduced in Sec. 5 of the current study. Results on additional simulations using bandwidth selector cerrd are reported in the Online Appendix B.
Table 4. Results of the simulations with the half-size bandwidth.
5. Multiple imputation as a graphical method of diagnosis for RDDs
Now that we are reasonably sure that MIRDDs can estimate the LATE at the cutoff, this article suggests to use MIRDDs as a diagnostic tool for RDDs. This is important, because graphical analyses are an integral part of RDD analyses (Imbens and Lemieux Citation2008, p. 622). As is shown in the simulations above, the selection of bandwidth is an important issue in using RDDs (Imbens and Lemieux Citation2008, p. 627). It is suggested to examine how the estimates of RDDs are sensitive to the changes in the model specification and bandwidth selection (Lee and Lemieux Citation2015, p. 312). Also, Gelman and Imbens (Citation2019, p. 448) argue that local low-order polynomials such as local linear or local quadratic models should be used in RDDs. The choice between these two models may be assessed using the diagnostic method presented in this section.
This section follows the spirit of Gelman et al. (Citation2005, p. 75), who proposed the predictive assessment to evaluate a model based on the predictions under available data. Using multiple imputation as a diagnostic tool is advantageous, because it allows graphical model checks that are easy to interpret without formally computing reference distributions (Gelman et al. Citation2005, p. 75). Indeed, in the context of missing data analysis by multiple imputation, graphical diagnostics of comparing observed and imputed data are now common (Abayomi, Gelman, and Levy Citation2008; Honaker, King, and Blackwell Citation2011, pp. 25–29; van Buuren Citation2018, pp. 190–194). To implement the methods described in this section, the author has written easy-to-use software, R-package MIRDD. See the Appendix about this software.
5.1. Procedure
Following the procedure explained in Gelman et al. (Citation2005, p. 77), the model check by MIRDDs in the current study proceeds in the following three steps.
Step 1: Construct multiply-imputed datasets, and compute a point estimate for the LATE at the cutoff point. This can be done in the same manner as in Sec. 3. In the software provided in the Appendix,
is set to 100 by default, but the user can freely change this number.
Step 2: Construct a graph using imputed data. The
sets of multiply-imputed data show
possibilities for the LATE at the cutoff point. Depending on the graph types, if the graphs are not overwhelmed by
sets of results, the software will display
results by default for the purpose of including estimation uncertainty. Otherwise, only one result will be shown. Note that Gelman et al. (Citation2005, p. 77) contend that displaying one result is good enough. Also, in the software provided in the Appendix, the user can freely change the number of imputed data to be displayed in a graph.
Step 3: Construct the reference distribution. In our case, this is simply the distribution of observed data.
5.2. Application to the incumbency advantage in U.S. House elections
For the purpose of illustration, we will apply the proposed method to the real data on the incumbency advantage in U.S. House elections, which was originally analyzed by Lee (Citation2008) using the sharp RDD. This dataset is publicly available in Olivares and Sarmiento-Barbieri (Citation2020, p. 5). As noted in Sec. 3.1, this is the real dataset based on which design 1 in the simulations was generated in EquationEq. (18)(18)
(18) . In this dataset, the forcing variable
is the difference in vote share between the Democratic and Republican parties in election at
and the dependent variable
is vote share in election at
In this example, we are interested in whether a candidate for the U.S. House of Representatives has an advantage if the candidate’s party won the seat in the previous election (Angrist and Pischke Citation2009, p. 257).
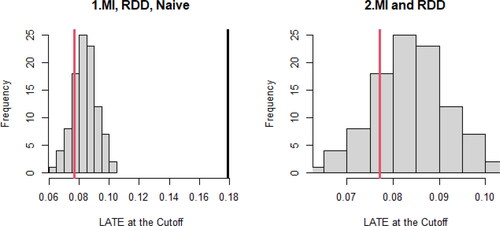
displays the results of the analyses, replicating the analyses in Lee (Citation2008). The estimates from MIRDD and RDD are 0.084 and 0.077, respectively, meaning that the estimated incumbency advantage is about 8% of the vote share (Lee Citation2008, p. 690). The 95% confidence intervals are and
respectively. Thus, we are 95% confident that the incumbency advantage at the cutoff point is roughly 6 to 10% of the vote share.
Table 5. Results of the analyses.
Note that, the bandwidth in is about twice as large as the automatically selected size by mserd. The simulations reported in and in the Online Appendix A showed that a narrower bandwidth tended to lead to better results. Readers can freely use the software in the Appendix to see how different settings may affect the conclusions we make. Below, we will diagnose whether the results in are reasonable or not.
graphically presents the results in . The solid black line is the Naïve estimator, the solid red line is RDD, and the histogram is MIRDD (). We can visually confirm that Naïve is far away from MIRDD and RDD, while RDD is within the distribution of MIRDD. When the conclusions from MIRDD and RDD agree, it still may not guarantee that the conclusion is correct, but we may feel more confident, because the disagreement between MIRDD and RDD indicates that at least one of them is wrong.
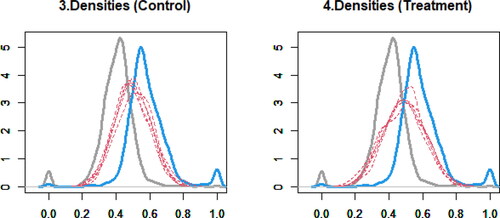
presents the densities of observed and imputed data in the control and treatment groups, where the gray solid curve is the density of observed data in the control group, the blue solid curve is the density of observed data in the treatment group, and the red dashed curves are the densities of imputed data. We look for anomalous shapes in imputed data compared to observed data. These figures show that the distributions of imputed data are consistent with the implications from observed data. Comparing the densities of observed and imputed data is a standard diagnostic technique in multiple imputation of missing data (Abayomi, Gelman, and Levy Citation2008, p. 277; Honaker, King, and Blackwell Citation2011, p. 26; van Buuren Citation2018, p. 193).
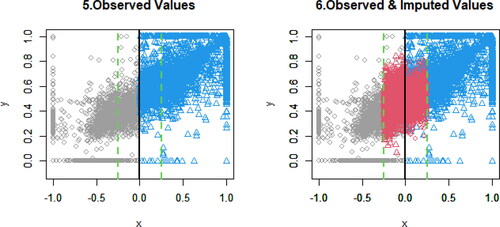
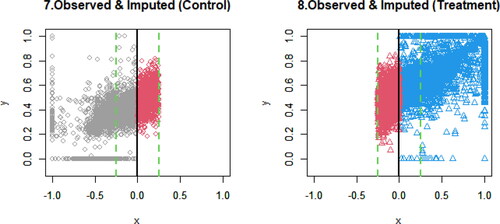
presents the scatterplots of observed and imputed data. The black solid line is the cutoff point. The green dashed lines are the bandwidths, where the RDD analysis is performed. Gray circles are the observed data in the control group, and blue triangles are the observed data in the treatment group. Red circles are the imputed data for the control group, and red triangles are the imputed data for the treatment group. The distributions of imputed data look consistent with those of observed data; however, the distributions of imputed data overlap those of observed data, making it somewhat hard to visualize.
To provide clearer pictures, divides the right-hand side panel in into two figures. In , the left-hand panel focuses on the control group, and the right-hand panel focuses on the treatment group. Beyond the cutoff point, red circles and red triangles are the extrapolation, respectively. Based on the implications from the observed data (gray circles and blue triangles), the extrapolation looks reasonable.
Figure 4. Left-hand panel is the scatterplot of observed and imputed data () in the control group, and right-hand panel is the scatterplot of observed and imputed data (
) in the treatment group.
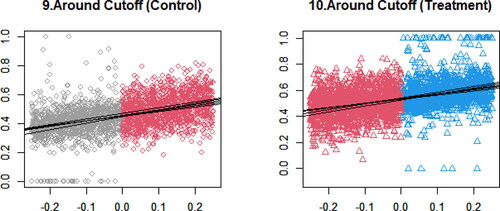
further magnifies , focusing on the local area chosen by the bandwidth. In this case, the horizontal axis ranges from −0.25 to 0.25. Five solid lines are the estimated local slopes based on imputed data. These figures confirm that local linearity approximately holds in this example.
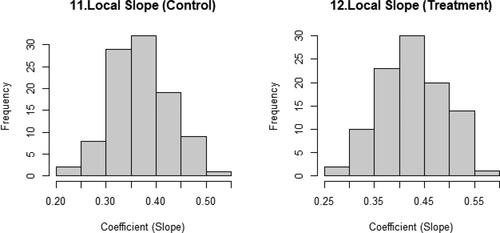
presents the histograms of the local slopes for the control and treatment groups. showed only five lines, so as to clearly show the scatterplot. displays the distribution of all of the local slopes. These figures show the estimation uncertainty in the local area, meaning that if the distributions are too wide or oddly shaped, we are less certain of the results.
6. Conclusion
The RDD is one of the strongest causal inference methods for non-experimental data, because RDDs focus on the small enough neighborhood around the cutoff point, so that subgroups can be treated as de-facto randomly assigned groups. Thus, RDDs can estimate the LATE at the cutoff point. This article found that the proposed method (MIRDDs) can also estimate the LATE at the cutoff point, just as equally well as RDDs. Based on this finding, this article proposed a graphical method based on MIRDDs to diagnose the validity of RDD analyses. This is an important addition to the literature of RDDs in particular and of causal inference in general, because graphical assessments are an integral part of RDD analyses. Furthermore, an easy-to-use software program is provided that implements the methods explained in this article.
Supplemental Material
Download MS Word (55.4 KB)Supplemental Material
Download MS Word (302.1 KB)References
- Abayomi, K., A. Gelman, and M. Levy. 2008. Diagnostics for multivariate imputations. Journal of the Royal Statistical Society: Series C (Applied Statistics) 57 (3):273–91. doi:10.1111/j.1467-9876.2007.00613.x.
- Allison, P. D. 2002. Missing data. Thousand Oaks: Sage Publications.
- Angrist, J. D., and J. S. Pischke. 2009. Mostly harmless econometrics: An empiricist’s companion. Princeton: Princeton University Press.
- Bodner, T. E. 2008. What improves with increased missing data imputations? Structural Equation Modeling: A Multidisciplinary Journal 15 (4):651–75. doi:10.1080/10705510802339072.
- Branson, Z., M. Rischard, L. Bornn, and L. W. Miratrix. 2019. A nonparametric Bayesian methodology for regression discontinuity designs. Journal of Statistical Planning and Inference 202:14–30. doi:10.1016/j.jspi.2019.01.003.
- Calonico, S., M. D. Cattaneo, and R. Titiunik. 2020. Package ‘rdrobust’. The Comprehensive R Archive Network. https://cran.r-project.org/web/packages/rdrobust/rdrobust.pdf. Accessed January 27, 2021.
- Calonico, S., M. D. Cattaneo, and R. Titiunik. 2014. Robust nonparametric confidence intervals for regression-discontinuity designs. Econometrica 82 (6):2295–326. doi:10.3982/ECTA11757.
- Carpenter, J. R., and M. G. Kenward. 2013. Multiple imputation and its application. Chichester, West Sussex: A John Wiley & Sons Publication.
- Carsey, T. M., and J. J. Harden. 2014. Monte Carlo simulation and resampling methods for social science. Thousand Oaks: Sage Publications.
- Dore, D. D., S. Swaminathan, R. Gutman, A. N. Trivedi, and V. Mor. 2013. Different analyses estimate different parameters of the effect of erythropoietin stimulating agents on survival in end stage renal disease: A comparison of payment policy analysis, instrumental variables, and multiple imputation of potential outcomes. Journal of Clinical Epidemiology 66 (8 Suppl):S42–S50. doi:10.1016/j.jclinepi.2013.02.014.
- Enders, C. K. 2010. Applied missing data analysis. New York: The Guilford Press.
- Gelman, A., and G. Imbens. 2019. Why high-order polynomials should not be used in regression discontinuity designs. Journal of Business & Economic Statistics 37 (3):447–56. doi:10.1080/07350015.2017.1366909.
- Gelman, A., I. V. Mechelen, G. Verbeke, D. F. Heitjan, and M. Meulders. 2005. Multiple imputation for model checking: Completed-data plots with missing and latent data. Biometrics 61 (1):74–85. doi:10.1111/j.0006-341X.2005.031010.x.
- Graham, J. W., A. E. Olchowski, and T. D. Gilreath. 2007. How many imputations are really needed? Some practical clarifications of multiple imputation theory. Prevention Science 8 (3):206–13. doi:10.1007/s11121-007-0070-9.
- Gujarati, D. N. 2003. Basic econometrics. 4th ed. Boston: McGraw-Hill.
- Gutman, R., and D. B. Rubin. 2013. Robust estimation of causal effects of binary treatments in unconfounded studies with dichotomous outcomes. Statistics in Medicine 32 (11):1795–814. doi:10.1002/sim.5627.
- Gutman, R., and D. B. Rubin. 2015. Estimation of causal effects of binary treatments in unconfounded studies. Statistics in Medicine 34 (26):3381–98. doi:10.1002/sim.6532.
- Hahn, J., P. Todd, and W. Klaauw. 2001. Identification and estimation of treatment effects with a regression-discontinuity design. Econometrica 69 (1):201–9. doi:10.1111/1468-0262.00183.
- Holland, P. W. 1986. Statistics and causal inference. Journal of the American Statistical Association 81 (396):945–60. doi:10.1080/01621459.1986.10478354.
- Holland, P. W., and D. B. Rubin. 1988. Causal inference in retrospective studies. Evaluation Review 12 (3):203–31. doi:10.1177/0193841X8801200301.
- Honaker, J., and G. King. 2010. What to do about missing values in time-series cross-section data. American Journal of Political Science 54 (2):561–81. doi:10.1111/j.1540-5907.2010.00447.x.
- Honaker, J., G. King, and M. Blackwell. 2011. Amelia II: A program for missing data. Journal of Statistical Software 45 (7):1–47. doi:10.18637/jss.v045.i07.
- Honaker, J., G. King, and M. Blackwell. 2019. Package ‘Amelia’. The Comprehensive R Archive Network. https://cran.r-project.org/web/packages/Amelia/Amelia.pdf. Accessed January 27, 2021.
- Horowitz, J. L. 2001. The bootstrap. In Handbook of econometrics, ed. J. J. Heckman and E. Leamer, Vol. 5, 3160–228. North Holland: Elsevier.
- Imbens, G. W., and D. B. Rubin. 2015. Causal inference for statistics, social, and biomedical sciences: An introduction. New York: Cambridge University Press.
- Imbens, G. W., and T. Lemieux. 2008. Regression discontinuity designs: A guide to practice. Journal of Econometrics 142 (2):615–35. doi:10.1016/j.jeconom.2007.05.001.
- Imbens, G., and K. Kalyanaraman. 2012. Optimal bandwidth choice for the regression discontinuity estimator. The Review of Economic Studies 79 (3):933–59. doi:10.1093/restud/rdr043.
- Kim, Y., and P. Steiner. 2016. Quasi-experimental designs for causal inference. Educational Psychologist 51 (3–4):395–405. doi:10.1080/00461520.2016.1207177.
- King, G., J. Honaker, A. Joseph, and K. Scheve. 2001. Analyzing incomplete political science data: An alternative algorithm for multiple imputation. American Political Science Review 95 (1):49–69. doi:10.1017/S0003055401000235.
- Lee, D. S. 2008. Randomized experiments from non-random selection in U.S. house elections. Journal of Econometrics 142 (2):675–97. doi:10.1016/j.jeconom.2007.05.004.
- Lee, D. S., and T. Lemieux. 2015. Regression discontinuity designs in social sciences. In The Sage handbook of regression analysis and causal inference, ed. H. Best and C. Wolf. Thousand Oaks: Sage Publications.
- Little, R. J. A., and D. B. Rubin. 2000. Causal effects in clinical and epidemiological studies via potential outcomes: Concepts and analytical approaches. Annual Review of Public Health 21:121–45. doi:10.1146/annurev.publhealth.21.1.121.
- Little, R. J. A., and D. B. Rubin. 2020. Statistical analysis with missing data. 3rd ed. Hoboken: John Wiley & Sons.
- Ludwig, J., and D. L. Miller. 2007. Does head start improve children’s life chances? Evidence from a regression discontinuity design. The Quarterly Journal of Economics 122 (1):159–208. doi:10.1162/qjec.122.1.159.
- Mooney, C. Z. 1997. Monte Carlo simulation. Thousand Oaks: Sage Publications.
- Nilsson, A., C. Bonander, U. Strömberg, and J. Björk. 2019. Assessing heterogeneous effects and their determinants via estimation of potential outcomes. European Journal of Epidemiology 34 (9):823–35. doi:10.1007/s10654-019-00551-0.
- Olivares, M., and I. Sarmiento-Barbieri. 2020. Package ‘RATest’. The Comprehensive R Archive Network. https://cran.r-project.org/web/packages/RATest/RATest.pdf. Accessed January 27, 2021.
- Olszewski, A. J., C. Chen, R. Gutman, S. P. Treon, and J. J. Castillo. 2017. Comparative outcomes of immunochemotherapy regimens in Waldenström macroglobulinaemia. British Journal of Haematology 179 (1):106–15. doi:10.1111/bjh.14828.
- Rubin, D. B. 1974. Estimating causal effects of treatments in randomized and nonrandomized studies. Journal of Educational Psychology 66 (5):688–701. doi:10.1037/h0037350.
- Rubin, D. B. 1987. Multiple imputation for nonresponse in surveys. New York, NY: John Wiley & Sons.
- Rubin, D. B. 2004. Direct and indirect causal effects via potential outcomes. Scandinavian Journal of Statistics 31 (2):161–70. doi:10.1111/j.1467-9469.2004.02-123.x.
- Schafer, J. L. 1997. Analysis of incomplete multivariate data. Boca Raton: Chapman & Hall/CRC.
- Shadish, W. R., T. D. Cook, and D. T. Campbell. 2002. Experimental and quasi-experimental designs for generalized causal inference. Boston: Houghton Mifflin Company.
- Stuart, E. A. 2010. Matching methods for causal inference: A review and a look forward. Statistical Science 25 (1):1–21. doi:10.1214/09-STS313.
- Takahashi, M. 2017. Statistical inference in missing data by MCMC and Non-MCMC multiple imputation algorithms: Assessing the effects of between-imputation iterations. Data Science Journal 16 (37):1–17. doi:10.5334/dsj-2017-037.
- Thoemmes, F., W. Liao, and Z. Jin. 2017. The analysis of the regression-discontinuity design in R. Journal of Educational and Behavioral Statistics 42 (3):341–60. doi:10.3102/1076998616680587.
- van Buuren, S. 2018. Flexible imputation of missing data. 2nd ed. Boca Raton, FL: Chapman & Hall/CRC.
- Westreich, D., J. K. Edwards, S. R. Cole, R. W. Platt, S. L. Mumford, and E. F. Schisterman. 2015. Imputation approaches for potential outcomes in causal inference. International Journal of Epidemiology 44 (5):1731–7. doi:10.1093/ije/dyv135.
- Winship, C., and S. L. Morgan. 1999. The estimation of causal effects from observational data. Annual Review of Sociology 25 (1):659–706. doi:10.1146/annurev.soc.25.1.659.
Appendix
To implement the approaches introduced in this article, the author has written easy-to-use software, R-package MIRDD. The software and documentation are freely available at https://github.com/mtakahashi123/MIRDD. Essentially, analysis can be easily done by just typing MIdiagRDD(y, x, cut) in R, with a variety of other options also available. Replication materials for the simulations are deposited at https://github.com/mtakahashi123/replication1.