Abstract
We report the discovery of a novel β-globin gene variant, Hb Odder, characterized by a single nucleotide substitution; HBB:c.316C > G; CD105 (Leu > Val). This variant emerged incidentally during routine HbA1c measurements for diabetes monitoring. The patient exhibited no clinical or biochemical evidence of anemia or hemolysis. Our data on this variant suggest that Hb Odder is benign, regrettably limitations in our data make formal evaluations of stability and oxygen affinity impossible; additionally this emphasizes the importance of considering hemoglobin variants in the differential diagnosis of abnormal Hb A1c levels and suggest that laboratories should use alternative methods for the correct measurement of Hb A1c when hemoglobin variants interfere with diabetes monitoring. Notably, three other mutations have been described at codon 105 of the β globin chains and correspond to three Hb variants with different characteristics: Hb South Milwaukee, Hb Bellevue IV and Hb St. George.
To date, more than 900 variations in the β-globin gene have been characterized [Citation1,Citation2]. Their impacts differ with many being asymptomatic, whereas others may cause thalassemia-like conditions, and some affect hemoglobin properties such as stability or oxygen affinity.
Patients are commonly tested for hemoglobinopathies as part of a screening program [Citation3] or when they present with clinical signs suggestive of a hemoglobinopathy, such as microcytic and hypochromic anemia.
It is important to note that although hemoglobin variants often do not cause clinical symptoms, they can interfere with the measurement of Hb A2 and even Hb A1c (glycated Hb A1). However, separative methods such as high-performance liquid chromatography (HPLC) or capillary electrophoresis (CE), which are widely used for hemoglobinopathy screening and diabetes screening and monitoring, may reveal the presence of Hb variants. In addition to allowing the random detection of asymptomatic Hb variants, they also suggest that the laboratory use alternative methods for the correct measurement of Hb A1c [Citation4].
In this paper, we discuss the case of a 36-year-old man who was found to have a novel β-globin variant. The variant was briefly mentioned in an article from 2018 reporting on hemoglobin variants found accidentally on Hb A1c testing [Citation5]. Despite his young age the patient had had multiannual measurements of Hb A1c, all but one showing values >48 mmol/mol, the diagnostic cutoff for diabetes (). Other routine blood samples performed were normal, although MCV and MCHC measurements were low, but within the reference range and just below the reference range respectively (). Initially, measurements of Hb A1c were carried out using HPLC () (Variant II Turbo, Bio-Rad, United States), which was inconspicuous. Later, analysis of Hb A1c was performed using CE (Capillarys 3 TERA, Sebia, France), which revealed the abnormal variant as an extra peak on the curve, denoted 4, as indicated with an arrow on . Notably, assessment of Hb A1c was unavailable due to the atypical profile. Due to the extra peak, diagnostic samples were sent for hemoglobin testing at the Danish Red Blood Cell Center (DRBC). HPLC was performed as previously described [Citation6] using Waters UPLC (Waters, Milford, MA, USA). The first hemoglobinopathy testing failed to detect the variant and the sample was categorized as normal. However, a repeated sample led to Sanger sequencing, using Applied Biosystems 3500 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA), of HBB which revealed a previously undescribed missense variant HBB:c.316C > G; CD105 (Leu > Val) (). We propose to call this variant, Hb Odder named after a city close to the patient’s origin. The two subjects compared in and are the same ones analyzed with two different methods: HPLC (Variant II Turbo, Bio-Rad, United States) and CE (Capillarys 3 TERA, Sebia, France) respectively. In panel A the patient with the Hb Odder variant and in panel B a normal control patient.
Figure 1. (A) Hb A1c measurement of the man carrying the novel variant using HPLC, not revealing the novel β-globin variant above. Below (B) HPLC evaluations of an individual not carrying any hemoglobin variants. Both evaluations were carried out using Variant II Turbo, Bio-Rad, United States.
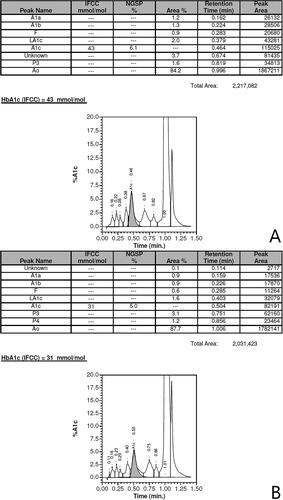
Figure 2. (A) Hb A1c measurement of the man carrying the novel variant using CE revealing the novel β-globin variant above. The peak revealing the abnormal variant is indicated with an arrow, notably the assessment of Hb A1C was unavailable due to the atypical profile. Below (B) CE evaluations of an individual not carrying any hemoglobin variants. Both evaluations were carried out using Capillarys 3 TERA, Sebia, France.
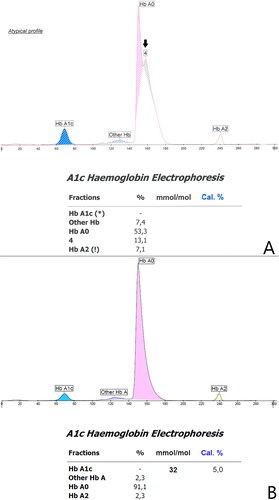
Figure 3. Sanger sequencing of HBB revealing the missense variant. The upper strand is the forward sequence, the lower the reverse sequence. Evaluations were carried out using Applied Biosystems 3500 Genetic Analyzer (Life Technologies, Carlsbad, CA).
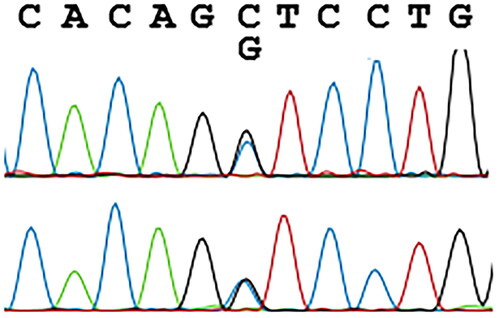
Table 1. Measurements of Hb A1c over a course of 3 years.
Table 2. Routine blood counts drawn 3 months apart with no signs of hemolysis or anemia, notably the MCV values are low within reference range and MCHC just below reference range.
The patient exhibited no clinical or biochemical evidence of anemia or hemolysis, although MCV and MCHC measurements were low which could indicate a slightly unstable hemoglobin variant. Accordingly, the patient was tested for α thalassemia variants, which were not present. To our regret, no other tests were performed, and the patient was unavailable for further testing. The presence of the hemoglobin variant and its interference with CE evaluations, suggests that for this patient, another method for assessing glycemic control in diabetes monitoring should be used. Two missense variants in HBB CD105 are listed in the HbVar and Ithanet databases: Hb South Milwaukee (Leu > Phe) and Hb Bellevue IV (Leu > Pro). Hb South Milwaukee was described in 1990 [Citation7] and is considered pathogenic due to its increased oxygen affinity and reduced cooperativity. Hb Bellevue IV, though not yet documented in a published paper, is reported to be unstable, but its clinical impact is unclear [Citation8]. Another variant, Hb St. George (Leu > His), is listed in neither database, but is reported to be pathogenic due to mild instability and decreased oxygen affinity, causing hypoxemia and mild anemia [Citation9].
Our data on this variant suggest that Hb Odder is benign. Regrettably, limitations in our data make formal evaluations of stability and oxygen affinity impossible. Additionally, they emphasize the importance of considering hemoglobin variants in the differential diagnosis of abnormal Hb A1c levels and suggest that laboratories use alternative methods for the correct measurement of Hb A1c when hemoglobin variants interfere with diabetes monitoring. We have reported our obtained data to the relevant database [Citation10], which hopefully will be supplemented by future studies of this novel variant.
Ethical approval
Data were stored and handled in accordance with permission from the Danish Data Protection Agency (10122009 HEH-L.HB). The patient consented to all diagnostic tests performed.
Acknowledgements
This project was carried out within the framework of European Reference Network on Rare Haematological Diseases (ERN-EuroBloodNet)—Project ID No 10108571. ERN-EuroBloodNet is partly co-funded by the European Union within the framework of the Fourth EU Health Programme.
Disclosure statement
AG has done consulting for Agios, Bristol Myers Squibb, Novartis, Novo Nordisk, Pharmacosmos and received research support from Agois, Bristol Myers Squibb, Novo Nordisk, Saniona, Sanofi.
Additional information
Funding
References
- Kountouris P, Lederer CW, Fanis P, et al. IthaGenes: an interactive database for haemoglobin variations and epidemiology. PLoS One. 2014;9(7):e103020. doi:10.1371/journal.pone.0103020.
- Giardine B, Borg J, Viennas E, et al. Updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Nucleic Acids Res. 2014;42(Database issue):D1063–D1069.
- Glenthøj A, Samson M, Toft N, et al. Det danske screeningsprogram for hæmoglobinopatier [The Danish screening programme for haemoglobinopathies]. Ugeskr Laeger. 2021;183(3):1–8.
- Bry L, Chen PC, Sacks DB. Effects of hemoglobin variants and chemically modified derivatives on assays for glycohemoglobin. Clin Chem. 2001;47(2):153–163. doi:10.1093/clinchem/47.2.153.
- Nybo J, Hansen AT, Petersen JB, et al. Hemoglobin variants found in relation to HbA1c testing: high occurrence of Hb Athens-Georgia in the Northern Jutland, Denmark. Clin Chem Lab Med. 2019;57(6):e108–10–e110. Vol. doi:10.1515/cclm-2018-0902.
- Nygaard M, Petersen J, Bjerrum OW. Haemoglobinopathia Ypsilanti - a rare, but important differential diagnosis to polycythaemia vera. Leuk Res Rep. 2013;2(2):86–88.
- Honig GR, Vida LN, Latorraca R, et al. Hb South Milwaukee. Am J Hematol. 1990;34(3):199–203. doi:10.1002/ajh.2830340308.
- HbVar: A database of Human Hemoglobin Variants and Thalassemias. Hb Bellevue IV., HbVar. ID: 2819 [Internet]. 2011. [cited 2024 Mar 28]. Available from: https://globin.bx.psu.edu/cgi-bin/hbvar/query_vars3?mode=output&display_format=page&i=2819.
- Meznarich JA, Rets A, Agarwal AM, et al. Novel, de novo, beta-globin variant with decreased oxygen affinity (HBB:c.317T > A, “Hemoglobin St. George”) in a healthy child with low oxygen saturations and anemia. Am J Hematol. 2021;96(12):E448–E450. Vol doi:10.1002/ajh.26356.
- HbVar: A database of Human Hemoglobin Variants and Thalassemias. Hb Odder, HbVar ID: 3294 [Internet]. 2024. [cited 2024 Mar 28]. Available from: https://globin.bx.psu.edu/cgi-bin/hbvar/query_vars3?mode=output&display_format=page&i=3294&.cgifields=histD.
