ABSTRACT
Introduction.
The phylogeography of Scorpidium revolvens (Sw. ex Anonymo) Rubers in northwestern Europe, primarily Scandinavia, was explored. Possibly because of human activity and atmospheric pollution, the species is sparser in the south than in the north, therefore its genetic diversity may be lowest in the south.
Methods.
Relationships within Scorpidium revolvens were evaluated in a network context, based on nuclear ITS and plastid rpl16 data. The regional diversity and haplotype composition of Sc. revolvens, Sc. cossonii (Schimp.) Hedenäs and Sarmentypnum exannulatum (Schimp.) Hedenäs were compared.
Key results.
When the sister species Scorpidium cossonii was used as an outgroup to determine polarity, the basalmost Sc. revolvens haplotype was found to be arctic, and the closest haplotypes to this were found in northern or montane specimens. From a basal grade, three lineages evolved, with differential north and south distribution. The haplotype diversity of Sc. revolvens was lowest in the south, and the haplotype composition of Sc. revolvens, Sc. cossonii and Sarmentypnum exannulatum in the south differed from that in other regions.
Conclusions.
A northern origin for Scorpidium revolvens in Scandinavia is suggested, in common with Sc. cossonii. Wider geographical sampling might confirm a northern origin for Scorpidium. Geographical distributions of the three Sc. revolvens lineages suggest partly separate colonisation routes. Compared with the north, Sc. revolvens in the sparsely populated south has lower haplotype diversity and different haplotype composition. The two other species investigated also had different haplotype composition, and further investigations might determine whether this represents a general pattern in Scandinavia.
Introduction
Among bryophytes, wetland species may have been the subject of the largest number of studies of genetic variation and diversity. The results of these studies have led to a better understanding of several processes, such as cryptic speciation, hybridisation, incomplete lineage sorting, and horizontal plastid gene transfer (Hedenäs and Eldenäs Citation2007; Natcheva and Cronberg Citation2007; Bączkiewicz et al. Citation2017; Hedenäs et al. Citation2021; Meleshko et al. Citation2021; Hedenäs Citation2022). They have also improved our understanding of the phylogeography of both peat mosses (Sphagnum) and so-called ‘brown mosses’ (Stenøien et al. Citation2010; Mikulášková et al. Citation2017; Hedenäs Citation2019; Manukjanová et al. Citation2020; Hedenäs et al. Citation2022).
Scorpidium cossonii (Schimp.) Hedenäs is one of the brown mosses that has been extensively studied (Hedenäs Citation1989; Hedenäs and Eldenäs Citation2008; Hedenäs Citation2009). This species grows in mineral-rich to calcareous fens (Kooijman and Hedenäs Citation1991; Graham et al. Citation2019), whereas Sc. revolvens (Sw. ex Anonymo) Rubers, the sister species of the Sc. cossonii lineage, grows in relatively mineral-poor fens. Both Sc. revolvens and Sc. cossonii are relatively common in middle and northern Scandinavia. In southern Scandinavia, however, only Sc. cossonii is frequent in many regions (see map in Hedenäs Citation1989).
It seems unlikely that the habitat of Scorpidium revolvens should be naturally much rarer than that of Sc. cossonii in the south, because non-calcareous regions dominate most of southern Scandinavia according to the online geological map provided by the Swedish Geological Survey (https://apps.sgu.se/kartvisare/kartvisare-berg-50-250-tusen.html; accessed 30 May 2023). Reasons for the discrepancy include the possibility that the more intensive agriculture and forestry, and the earlier atmospheric deposition of S and N compounds, especially in the south, affected poorly buffered mires more strongly than calcareous fens, or that the former type of fen was drained more often (e.g. Naturvårdsverket Citation2007, Citation2022). The poorly buffered and sensitive fen habitat where Sc. revolvens grows could therefore have to a high extent been lost in the south (Kooijman Citation2012; Kolari et al. Citation2021). Whatever the reason for the relative rarity of Sc. revolvens in southern Scandinavia, this could potentially result in a lower genetic diversity in the southern compared with the northern portion of its distribution range (Frankham Citation1996; Leimu et al. Citation2006).
The present study was carried out to answer the following questions concerning Scorpidium revolvens in Scandinavia: (i) What are the phylogeographical patterns within this species?, and (ii) Is its genetic diversity in the more sparsely populated southern region lower than that farther north? Additionally, because preliminary results suggested that the genetic diversity of Sc. revolvens is indeed lower in the south, the regional diversity pattern of this species was compared with that of two other wetland mosses, focusing on three geographical regions for which sufficient data exist for all three species.
Material and methods
Studied material
Scorpidium revolvens is a medium-sized to robust pleurocarpous fen moss that often becomes purplish to blackish red when it grows exposed to sunshine. Scorpidium cossonii was not distinguished from Sc. revolvens in much of the earlier literature, and Sc. revolvens was frequently, but erroneously, considered to grow in mineral-rich fens. However, in contrast to Sc. cossonii, Sc. revolvens grows in fens of intermediate mineral richness (Kooijman and Hedenäs Citation1991; Graham et al. Citation2019). It also differs from Sc. cossonii in having a double chromosome number, being autoicous rather than dioicous, and having longer median stem leaf cells with more longly fusiformly narrowed ends. Additional distinguishing features of Sc. revolvens have been discussed by Hedenäs (Citation1989). It frequently reproduces sexually, producing easily dispersed small spores (13.5–21.0 μm; Hedenäs Citation1989). Scorpidium revolvens is widespread and often common in (northern) temperate to subpolar areas of the northern hemisphere, whereas it is infrequent or mostly rare in the southern hemisphere (Hedenäs Citation2003a, Citation2003b).
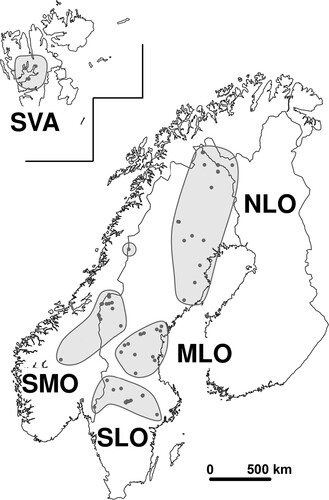
The intraspecific patterns within Scorpidium revolvens were explored based on 63 specimens. Four specimens represent Svalbard (SVA); 16, the northern Scandinavian lowland (NLO); 15, the southern Scandinavian mountains (SMO); 15, the middle Scandinavian lowland (MLO); and 13, the southern Scandinavian lowland (SLO). For the delimitations of these five regions, see . Except in the cases of specimens from outside Sweden, these regions correspond approximately with the lowlands of northern Sweden, the mountains of middle Sweden, the lowlands of middle Sweden, and southern Sweden in Hedenäs (Citation2019). Additionally, a single specimen from the middle of the mountain range was included. Scorpidium cossonii was chosen as an outgroup, based on the findings of Hedenäs and Eldenäs (Citation2008). The studied specimens are listed in Appendix 1.
Figure 1. Circumscription of the five Scandinavian regions that were compared for Scorpidium revolvens. MLO, middle Scandinavian lowland (15 specimens); NLO, northern Scandinavian lowland (16 specimens); SLO, southern Scandinavian lowland (13 specimens); SMO, southern Scandinavian mountains (15 specimens); SVA, Svalbard (4 specimens).
Sequence data for comparison with the rich-fen species Scorpidium cossonii and a species that grows in a habitat similar to that of Sc. revolvens but is more frequent throughout its range, namely Sarmentypnum exannulatum (Schimp.) Hedenäs, were extracted from Hedenäs (Citation2019), with enough specimens being available from the Sc. revolvens regions NLO (23 and 37 specimens, respectively), SMO (22 and 14, respectively) and SLO (12 and 20, respectively) (Appendix 2).
Molecular methods
Nuclear ITS and plastid rpl16 sequences were generated for the Scorpidium revolvens specimens, as described by Hedenäs (Citation2009), except as follows. The amplified PCR products were purified from excess primers and nucleotides by using ExoSap-IT (Applied Biosystems, Waltham, MA, USA). For all specimens, 5 μL of ExoSap-IT was added to 20 μL of PCR product and incubated at 37°C for 30 min; this was followed by an enzyme inactivation step at 80°C for 15 min. The purified PCR products, together with the same primers used for PCR amplification, were subsequently sent to Macrogen Europe (Amsterdam, The Netherlands) for single-stranded sequencing using the Applied Biosystems 3730XL sequencer.
Sequence editing and analysis
The nucleotide sequence fragments for each DNA region were edited and assembled using PhyDE 0.9971 (http://www.phyde.de/index.html; accessed 25 May 2023). The assembled sequences were aligned manually in PhyDE. Regions of partially incomplete data at the beginning and end of the sequences were identified and excluded from subsequent analyses. Gaps were coded using the simple indel coding of Simmons and Ochoterena (Citation2000) in SeqState (Müller Citation2005). Gaps provided additional information, which was included in the analyses. The sequence alignments used in the analyses are available as Supplemental material 1. GenBank accession numbers for the specimens are listed in Appendix 1.
ITS paralogues are occasionally encountered in bryophytes (Košnar et al. Citation2012; Hedenäs et al. Citation2019); however, the ITS chromatograms included in the present study did not show ‘messy’ patterns or noise that could suggest paralogy, and the 5.8S gene was invariable among specimens (Shaw et al. Citation2002; Feliner and Rosselló Citation2007). The revealed ITS variation was thus interpreted as being among homologous haplotypes.
A preliminary analysis did not reveal well-supported incongruences between ITS and rpl16 for the included specimens, and all molecular data were thus primarily analysed in combination to explore relationships. Initial analyses by TCS (Clement et al. Citation2000) and Neighbor-Net split networks in SplitsTree 4.12.6 (Huson and Bryant Citation2006) revealed reticulation, and the relationships were therefore analysed in a network context. Potential support for lineages in a tree context was evaluated by jackknife analyses (1000 replications) carried out using the program TNT (Goloboff et al. Citation2003).
The program TCS was used to identify haplotypes, based on ITS and rpl16 data separately. To investigate patterns of haplotype variation among the regions (see ), an analysis of molecular variance (Amova) was performed using GENALEX 6.501 (Peakall and Smouse Citation2006, Citation2012), with nuclear and plastid markers as separate loci. Pairwise ΦPT (an analogue of FST, i.e. genetic divergence among populations) was estimated using GENALEX, and the same program was used to calculate the effective number of haplotypes (Ne), Shannon’s information index (I; sometimes called Shannon’s diversity index), and the haplotype diversity (H) for each region. Differences in I between the regions were calculated using Hutcheson’s t-test (Hutcheson Citation1970; see also https://www.dataanalytics.org.uk/comparing-diversity/, accessed 21 February 2023).
Arlequin version 3.5.1.3 (Excoffier and Lischer Citation2010) was used to calculate nucleotide diversity (π), the average number of pairwise differences among regions, and Tajima’s D. Tajima’s D test of selective neutrality was used to decide whether the regional populations are probably stable in size, expanding or decreasing (Tajima Citation1989). Tajima’s D test was preferred over Fu’s FS test (Fu Citation1997), because recombination levels are unknown (Ramírez-Soriano et al. Citation2008). The null hypothesis in these analyses is that no differences exist in haplotype and nucleotide diversity or composition among the studied regions.
For both Scorpidium cossonii and Sarmentypnum exannulatum, an Amova was run to explore patterns of haplotype variation among the three regions and pairwise ΦPT, and Ne, I and H were calculated for the three regions. Differences in I between the regions were evaluated by Hutcheson’s t-test. These calculations were done as described above.
Results
The total number of aligned sites in the studied 63 specimens of Scorpidium revolvens, after deletion of regions at the beginnings and ends that were incomplete for some specimens, was 648 for ITS, including 7 base substitutions, of which 6 were parsimony-informative, and 1 indel, which was informative, and 619 for rpl16 (0 base substitutions; 3 indels, all parsimony-informative). Sequence lengths were 647–648 bp for ITS and 619–622 bp for rpl16.
Because no well-supported contradictions were found between ITS and rpl16, differences between the results based on these two markers individually and the concatenated dataset are mentioned in the text only when relevant. Based on the results for ITS and rpl16 together, both the Neighbor-Net split network and the TCS network grouped the Scorpidium revolvens specimens in a grade A and the somewhat indistinctly delimited lineages B–D (, 3). Only lineage C received moderate jackknife support (86%). This support comes from the ITS data; rpl16 data alone did not provide jackknife support for any of the lineages (results not shown).
Figure 2. Neighbor-Net split network for Scorpidium revolvens, based on data for the nuclear ITS and plastid rpl16, using one specimen of Sc. cossonii as an outgroup. Jackknife support of at least 80% is indicated by transverse grey lines. LyL, Lycksele lappmark; MLO, middle Scandinavian lowland; NLO, northern Scandinavian lowland; SLO, southern Scandinavian lowland; SMO, southern Scandinavian mountains; SVA, Svalbard.
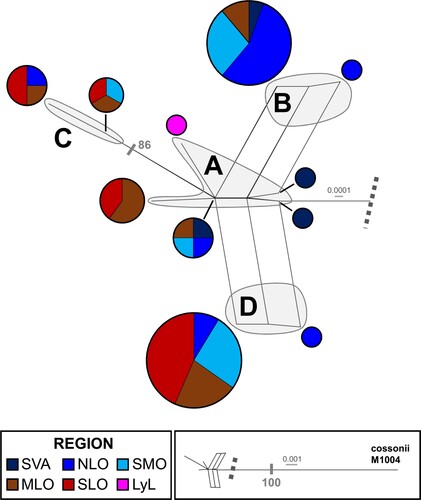
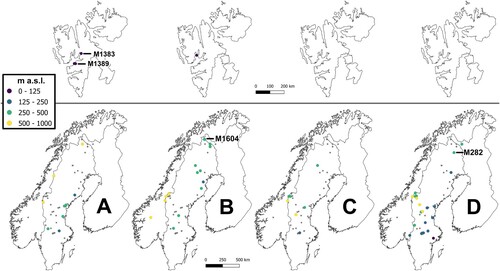
The geographical distributions of the four groups overlap in Scandinavia (), where lineage B seems to be relatively more common in the northern lowland region (NLO; see ) and southern mountainous Scandinavia (SMO), including one specimen from Svalbard (SVA). Lineages C and D are relatively more common in southern and middle Scandinavia (SLO and MLO, respectively), including SMO. Grade A shows a more even distribution in the sampled area, and includes three SVA specimens. The most basal specimen in the networks comes from Svalbard (specimen M1383; see , ). Overall, the specimens closest to the outgroup Scorpidium cossonii in the network were mainly collected in SVA, NLO or SMO. Specimens M282 and M1604, whose positions were suggested to be close to M1383 and M1389, respectively (see ), based on the data for rpl16 but not ITS (results not shown), were collected in northernmost Scandinavia (see , ).
Figure 3. TCS haplotype network for Scorpidium revolvens, based on data for the nuclear ITS and plastid rpl16 combined, using one specimen of Sc. cossonii as an outgroup. Specimens are indicated according to their numbers in Appendix 1. Black dots indicate ‘missing haplotypes’, that is, potential ‘haplotypes’ that were not recovered in the sampling.
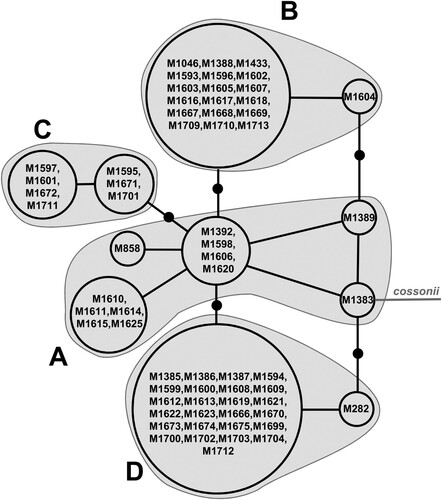
Figure 4. Geographical distributions of the specimens belonging to Scorpidium revolvens grade A (A) and lineages B–D (B, C, D) in and . Small grey dots indicate all sampled sites, whereas larger coloured dots indicate sites from which the respective groups were sampled. Different colours indicate different elevation spans. The Scandinavian specimens closest to the base of the network in (A) (cf. , 3) and two specimens in (B) and (D), which are discussed in the text, are indicated with lines and specimen numbers.
The Amova results revealed that 17% of the haplotype variation was among the regions, whereas 83% was within the regions (n = 63; ITS and rpl16, 5 + 4 haplotypes; Φ = 0.165, p = 0.006, 9999 permutations). A pairwise comparison of ΦPT values and average numbers of pairwise nucleotide differences showed that the composition of SVA differed from that of all other regions (haplotype data only); the composition of SLO differed from that of SVA, NLO and SMO; and the composition of NLO and MLO differed from each other (, A). Diversity was lowest in SVA (ITS data only) and SLO, significantly so in SLO compared with MLO for ITS and compared with the other three regions for rpl16, according to Shannon’s information index (). Tajima’s D test of selective neutrality did not suggest that the regional populations were increasing or decreasing (results not shown).
Figure 5. (A) Haplotype compositions in five regions for Scorpidium revolvens, plus a single specimen from Lycksele lappmark, based on data for either the nuclear ITS or the plastid rpl16 (total, n = 63). (B) Haplotype compositions in three regions for Sc. cossonii, based on combined data for the nuclear ITS or the plastid rpl16 (total, n = 57). (C) Haplotype compositions in three regions for Sarmentypnum exannulatum, based on data for the nuclear ITS or the plastid markers (rpl16, trnG, trnL–trnF) (total, n = 71). Circle sizes are proportional to the number of specimens in a region. The plastid marker haplotype compositions are indicated by green text and green circles around the pie charts. MLO, middle Scandinavian lowland; NLO, northern Scandinavian lowland; SLO, southern Scandinavian lowland; SMO, southern Scandinavian mountains; SVA, Svalbard.
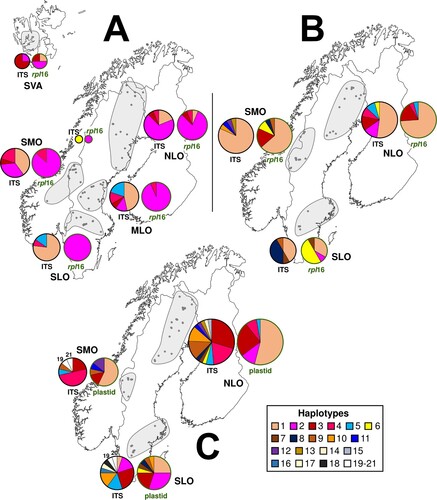
Table 1. Matrix of regions pairwise ΦPT values (A) and corrected average numbers of pairwise nucleotide differences (B) for ITS and rpl16 for the five regional populations of Scorpidium revolvens.Table Footnotea
Table 2. Haplotype and nucleotide diversity for the five studied regions for Scorpidium revolvens, based on data for ITS and rpl16.Table Footnotea
The Amova results showed that 11% (n = 70; ITS and rpl16, 13 + 9 haplotypes; Φ = 0.109, p = 0.004, 9999 permutations) or 4% (n = 81; ITS and plastid markers, 21 + 12 haplotypes; Φ = 0.042, p = 0.228, 9999 permutations) of the haplotype variation was among the three regions for Scorpidium cossonii and Sarmentypnum exannulatum, respectively. Pairwise comparison of the regions revealed significant differences in genetic composition between SLO and the other two regions for both these species (, B,C). Haplotype diversity was similar for all three regions ().
Table 3. Matrix of regions pairwise ΦPT values for three regions, with at least 10 specimens for Scorpidium cossonii, based on data for ITS and rpl16 (A), and for Sarmentypnum exannulatum, based on data for ITS and the combined data for plastid rpl16, trnG and trnL–trnF (B).Table Footnotea
Table 4. Haplotype diversity for three regions with at least 10 specimens for Scorpidium cossonii (13 ITS and 9 rpl16 haplotypes) and Sarmentypnum exannulatum (21 ITS and 12 plastid [rpl16, trnG and trnL–trnF] haplotypes).Table Footnotea
Discussion
Within Scorpidium revolvens, a basal grade gave rise to three different lineages with partly different geographical distributions in northwestern Europe. The basalmost haplotypes were collected in Svalbard, and numerous specimens of the basal grade were northern. In Scandinavia, the lowest genetic diversity was found in the southernmost region.
Intraspecific relationships in northwestern European Sc. revolvens
As has been found for its sister species Scorpidium cossonii (Hedenäs Citation2009, Citation2019), which was used as outgroup to determine the polarity within Sc. revolvens, the present results based on data for Scandinavia suggest that the most basal specimens of Sc. revolvens within grade A (see , 3) come from areas with cold climates. These two species are also similar in that members of the main lineages that originated from the basal grade occur in more temperate habitats. I tentatively suggest that ancestral haplotypes of Sc. revolvens are northern, and that the origin of the entire Scorpidium lineage could thus be northern. A wider geographical sampling of Sc. revolvens should be attempted to confirm or reject this hypothesis. Interestingly, early Pleistocene finds of Hamatocaulis vernicosus (Mitt.) Hedenäs, a member of the probable sister genus of Scorpidium (Hedenäs and Eldenäs Citation2008; Schlesak et al. Citation2018; Kučera et al. Citation2019), from northeastern and northwestern Greenland show that this species also occurred in the far north during earlier times (Bennike et al. Citation2022; Hedenäs and Bennike Citation2008).
Lineage B is, relatively, most frequent in the north and in the mountains, whereas the other two lineages (C and D) are more frequent in the southern portions of the sampled area. These differences could be a result of colonisation from different glacial refugia, as was suggested for Drepanocladus turgescens (T.Jensen) Broth., Rhytidium rugosum (Hedw.) Kindb., and to some degree, Dryas octopetala L. (Skrede et al. Citation2006; Hedenäs Citation2014, Citation2015; Hedenäs and Bisang Citation2019). It could also be a result of differences in climate adaptations, as has been suggested for different genetic entities of Pinus sylvestris L., Scorpidium cossonii and Timmia austriaca Hedw. (Eriksson et al. Citation1980; Hedenäs Citation2009, Citation2019, Citation2021).
Diversity patterns in northwestern European Sc. revolvens
A comparison with other studies of bryophytes shows that a relatively large proportion of the haplotype variation in Scorpidium revolvens is between regions (Hedenäs Citation2015, Citation2019). The most salient pattern is that the haplotype composition of the southern region, where the species occurs relatively sparsely, differs from most other regions, and that the diversity is clearly lowest in the south. For Sc. cossonii and Sarmentypnum exannulatum, a relatively large and small proportion of the haplotype variation, respectively, was between regions. No differences in diversity were found between the regions, but in both species, SLO differed significantly from NLO and SMO in terms of haplotype composition.
In moss species that are relatively frequent in northern Scandinavia, southern disjunct regional populations seem to differ markedly in genetic composition from those in regions farther north, a finding that suggests different postglacial origins, as in Drepanocladus turgescens and Rhytidium rugosum (Hedenäs Citation2014, Citation2015). The patterns seen in Scorpidium cossonii, Sc. revolvens and Sarmentypnum exannulatum suggest that in species with more continuous distributions, the southern regional populations may also have partly different origins than the regional populations of the north. Also, in flowering plants with wide distributions, we find genetic traces of different postglacial immigration routes, for example in Melica nutans L. (Tyler Citation2002) and Parnassia palustris L. (Borgen and Hultgård Citation2003). Fen habitats in southern Scandinavia are strongly affected by human management and atmospheric pollution (Naturvårdsverket Citation2007, Citation2022). The different genetic compositions found in southern regional populations of widespread fen mosses therefore suggest that southern populations may require conservation attention, even if such populations do presently not show signs of population decrease.
Supplemental Material 1
Download Text (84.2 KB)Acknowledgements
I thank Bodil Cronholm and Daniel Marquina for their efficient laboratory assistance, and Eva Mikulášková for providing three of the studied specimens. I appreciate the constructive comments from one reviewer and the handling editor. The newly sampled material comes from areas where genetic resources are free for non-commercial research.
Disclosure statement
No potential conflict of interest was reported by the author.
Supplemental material
Supplemental material for this article can be accessed here: https://doi.org/10.1080/03736687/2024.2318161.
Supplemental material 1. Alignment of ITS and rpl16.
Additional information
Funding
Notes on contributors
Lars Hedenäs
Lars Hedenäs has interests in bryophyte diversity, evolution, phylogeny and phylogeography. Wetland pleurocarps are a continuous theme in his research, but other pleurocarps as well as acrocarpous mosses are also among his research interests.
References
- Bączkiewicz A, Szczecińska M, Sawicki J, Stebel A, Buczkowska K. 2017. DNA barcoding, ecology and geography of the cryptic species of Aneura pinguis and their relationships with Aneura maxima and Aneura mirabilis (Metzgeriales, Marchantiophyta). PLOS One. 12(12):e0188837. https://doi.org/10.1371/journal.pone.0188837.
- Bennike O, Colgan W, Hedenäs L, Heiri O, Lemdahl G, Wiberg-Larsen P, Ribeiro S, Pronzato R, Manconi R, Bjørk AA. 2022. An Early Pleistocene interglacial deposit at Pingorsuit, North-West Greenland. Boreas. 52(1):27–41. https://doi.org/10.1111/bor.12596.
- Borgen L, Hultgård UM. 2003. Parnassia palustris: a genetically diverse species in Scandinavia. Botanical Journal of the Linnean Society. 142(4):347–372. https://doi.org/10.1046/j.1095-8339.2003.00186.x.
- Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 9(10):1657–1659. https://doi.org/10.1046/j.1365-294x.2000.01020.x.
- Eriksson G, Andersson S, Eiche V, Ifver J, Persson A. 1980. Severity index and transfer effects on survival and volume production of Pinus sylvestris in northern Sweden. Studia Forestalia Suecica. 156:1–32. https://pub.epsilon.slu.se/5442/1/SFS156.pdf.
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 10(3):564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x.
- Feliner GN, Rosselló JA. 2007. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution. 44(2):911–919. https://doi.org/10.1016/j.ympev.2007.01.013.
- Frankham R. 1996. Relationship of genetic variation to population size in wildlife. Conservation Biology. 10(6):1500–1508. https://doi.org/10.1046/j.1523-1739.1996.10061500.x. https://jstor.org/stable/2387021.
- Fu Y. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 147(2):915–925. https://doi.org/10.1093/genetics/147.2.915.
- Goloboff P, Farris J, Nixon K. 2003. Tree analysis using new technology. [accessed 2017 May 3]. http://www.lillo.org.ar/phylogeny/tnt/.
- Graham J, Farr G, Hedenäs L, Devez A, Watts MJ. 2019. Using water chemistry to define ecological preferences within the moss genus Scorpidium, from Wales, UK. Journal of Bryology. 41(3):197–204. https://doi.org/10.1080/03736687.2019.1603416.
- Hedenäs L. 1989. The genera Scorpidium and Hamatocaulis, gen. nov., in Northern Europe. Lindbergia. 15(1):8–36.
- Hedenäs L. 2003a. Amblystegiaceae (Musci). Flora Neotropica Monograph. 89:i–iv, 1–107.
- Hedenäs L. 2003b. The European species of the Calliergon–Scorpidium–Drepanocladus complex, including some related or similar species. Meylania. 28:1–116. https://www.bryolich.ch/pdfs/Meylania_ganze_Hefte/Meylania%2028.pdf.
- Hedenäs L. 2009. Relationships among Arctic and non-Arctic haplotypes of the moss species Scorpidium cossonii and Scorpidium scorpioides (Calliergonaceae). Plant Systematics and Evolution. 277:217–231. https://doi.org/10.1007/s00606-008-0131-y.
- Hedenäs L. 2014. Intraspecific genetic variation in selected mosses of Scandinavian interglacial refugia suggests contrasting distribution history patterns. Botanical Journal of the Linnean Society. 176(3):295–310. https://doi.org/10.1111/boj.12210.
- Hedenäs L. 2015. Rhytidium rugosum (Bryophyta) colonized Scandinavia from at least two glacial refugial source populations. Botanical Journal of the Linnean Society. 179(4):635–657. https://doi.org/10.1111/boj.12341.
- Hedenäs L. 2019. On the frequency of northern and mountain genetic variants of widespread species: essential biodiversity information in a warmer world. Botanical Journal of the Linnean Society. 191(4):440–474. https://doi.org/10.1093/botlinnean/boz061.
- Hedenäs L. 2021. Relationships within Timmia, especially within T. austriaca Hedw. (Musci, Timmiaceae). Journal of Bryology. 43(3):283–291. https://doi.org/10.1080/03736687.2021.1963914.
- Hedenäs L. 2022. Potentially misleading phylogenetic signal and its explanation in single species studies – contrasting Loeskypnum badium, Sarmentypnum exannulatum and Warnstorfia fluitans. The Bryologist. 125(1):102–114. https://doi.org/10.1639/0007-2745-125.1.102.
- Hedenäs L, Bennike O. 2008. A Plio-Pleistocene moss assemblage from Store Koldewey, NE Greenland. Lindbergia. 33(1):23–37. https://www.jstor.org/stable/27809535.
- Hedenäs L, Bisang I. 2019. Are the remains of the Central European population of Drepanocladus turgescens genetically distinct from Scandinavian populations? Herzogia. 32(1):209–218. https://doi.org/10.13158/heia.32.1.2019.209.
- Hedenäs L, Collart F, Heras P, Infante M, Kooijman A, Kučera J. 2022. Distributions and habitats of the two partly allopatric cryptic species of the vulnerable moss Hamatocaulis vernicosus (Bryophyta) in Europe. Botanical Journal of the Linnean Society. 200(2):233–254. https://doi.org/10.1093/botlinnean/boac011.
- Hedenäs L, Eldenäs P. 2007. Cryptic speciation, habitat differentiation, and geography in Hamatocaulis vernicosus (Calliergonaceae, Bryophyta). Plant Systematics and Evolution. 268:131–145. https://doi.org/10.1007/s00606-007-0529-y.
- Hedenäs L, Eldenäs P. 2008. Relationships in Scorpidium (Calliergonaceae, Bryophyta), especially between S. cossonii and S. scorpioides. Taxon. 57(1):121–130. https://www.jstor.org/stable/25065953.
- Hedenäs L, Heinrichs J, Gallego MT. 2019. The Scandinavian Syntrichia ruralis complex (Musci, Pottiaceae): a chaos of diversification. Plant Systematics and Evolution. 305:639–661. https://doi.org/10.1007/s00606-019-01596-0.
- Hedenäs L, Larsson P, Cronholm B, Bisang I. 2021. Evidence of horizontal gene transfer between land plant plastids has surprising conservation implications. Annals of Botany. 127(7):903–908. https://doi.org/10.1093/aob/mcab021.
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 23(2):254–267. https://doi.org/10.1093/molbev/msj030.
- Hutcheson K. 1970. A test for comparing diversities based on the Shannon formula. Journal of Theoretical Biology. 29(1):151–154. https://doi.org/10.1016/0022-5193(70)90124-4.
- Kolari THM, Korpelainen P, Kumpula T, Tahvanainen T. 2021. Accelerated vegetation succession but no hydrological change in a boreal fen during 20 years of recent climate change. Ecology and Evolution. 11(12):7602–7621. https://doi.org/10.1002/ece3.7592.
- Kooijman AM. 2012. ‘Poor rich fen mosses’: atmospheric N-deposition and P-eutrophication in base-rich fens. Lindbergia. 35:42–52. https://www.jstor.org/stable/lindbergia.35.42.
- Kooijman AM, Hedenäs L. 1991. Differentiation in habitat requirements within the genus Scorpidium, especially between S. revolvens and S. cossonii. Journal of Bryology. 16(4):619–627. https://doi.org/10.1179/jbr.1991.16.4.619.
- Košnar J, Herbstová M, Kolář F, Koutecký P, Kučera J. 2012. A case study of intragenomic ITS variation in bryophytes: assessment of gene flow and role of polyploidy in the origin of European taxa of the Tortula muralis (Musci: Pottiaceae) complex. Taxon. 61(4):709–720. https://doi.org/10.1002/tax.614001.
- Kučera J, Kuznetsova OI, Manukjanová A, Ignatov MS. 2019. A phylogenetic revision of the genus Hypnum: towards completion. Taxon. 68(4):628–660. https://doi.org/10.1002/tax.12095.
- Leimu R, Mutikainen P, Koricheva J, Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation? Journal of Ecology. 94(5):942–952. https://doi.org/10.1111/j.1365-2745.2006.01150.x.
- Manukjanová A, Košnar J, Kučera J. 2020. Genetic variation in two cryptic species of the rare fen moss Hamatocaulis vernicosus in the Czech Republic. Preslia. 92(1):57–72. https://doi.org/10.23855/preslia.2020.057.
- Meleshko O, Martin MD, Korneliussen TS, Schröck C, Lamkowski P, Schmutz J, Healey A, Piatkowski BT, Shaw AJ, Weston DJ, et al. 2021. Extensive genome-wide phylogenetic discordance is due to incomplete lineage sorting and not ongoing introgression in a rapidly radiated bryophyte genus. Molecular Biology and Evolution. 38(7):2750–2766. https://doi.org/10.1093/molbev/msab063.
- Mikulášková E, Veleba A, Šmerda J, Knoll A, Hájek M. 2017. Microsatellite variation in three calcium-tolerant species of peat moss detected specific genotypes of Sphagnum warnstorfii on magnesium-rich bedrock. Preslia. 89(2):101–114. https://doi.org/10.23855/preslia.2017.101.
- Müller K. 2005. SeqState. Applied Bioinformatics. 4:65–69. https://doi.org/10.2165/00822942-200504010-00008.
- Natcheva R, Cronberg N. 2007. Recombination and introgression of nuclear and chloroplast genomes between the peat mosses, Sphagnum capillifolium and Sphagnum quinquefarium. Molecular Ecology. 16(4):811–818. https://doi.org/10.1111/j.1365-294X.2006.03163.x.
- Naturvårdsverket. 2007. Myllrande våtmarker. Underlagsrapport till fördjupad utvärdering av miljömålsarbetet. Naturvårdsverket Rapport. 5771:1–132.
- Naturvårdsverket. 2022. Bara naturlig försurning. Fördjupad utvärdering av miljömålen 2023. Naturvårdsverket Rapport. 7069:1–73.
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x.
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics. 28(19):2537–2539. https://doi.org/10.1093/bioinformatics/bts460.
- Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A. 2008. Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics. 179(1):555–567. https://doi.org/10.1534/genetics.107.083006.
- Schlesak S, Hedenäs L, Nebel M, Quandt D. 2018. Cleaning a taxonomic dustbin: placing the European Hypnum species in a phylogenetic context! Bryophyte Diversity and Evolution. 40(2):37–54. https://doi.org/10.11646/bde.40.2.3.
- Shaw AJ, McDaniel SF, Werner O, Ros RM. 2002. New frontiers in bryology and lichenology. Phylogeography and phylodemography. The Bryologist. 105(3):373–383. https://doi.org/10.1639/0007-2745(2002)105[0373:PAP]2.0.CO;2.
- Simmons MP, Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 49(2):369–381. https://doi.org/10.1093/sysbio/49.2.369.
- Skrede I, Eidensen PB, Portela RP, Brochmann C. 2006. Refugia, differentiation and postglacial migration in arctic-alpine Eurasia, exemplified by the mountain avens (Dryas octopetala L.). Molecular Ecology. 15(7):1827–1840. https://doi.org/10.1111/j.1365-294X.2006.02908.x.
- Stenøien HK, Shaw AJ, Shaw B, Hassel K, Gunnarsson U. 2010. North American origin and recent European establishments of the Amphi-Atlantic peat moss Sphagnum angermanicum. Evolution. 65(4):1181–1194. https://doi.org/10.1111/j.1558-5646.2010.01191.x.
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123(3):585–595. https://doi.org/10.1093/genetics/123.3.585.
- Tyler T. 2002. Large-scale geographic patterns of genetic variation in Melica nutans, a widespread Eurasian woodland grass. Plant Systematics and Evolution. 236:73–87. https://doi.org/10.1007/s00606-002-0235-8.
Appendix
Appendix 1. Specimen data and GenBank accession numbers for specimens of Scorpidium revolvens. Data format: Specimen no. (grade/lineage [cf. Figures 2, 3]: haplotype ITS, haplotype rpl16): Locality; Collection year, Collector (collector’s no. [LH = L.Hedenäs]); Herbarium: registration no.; GenBank accession numbers for ITS, rpl16. Newly generated sequences have GenBank accession numbers beginning with ‘ON’.
Scorpidium revolvens (Sw. ex Anonymo) Rubers: M282 (D:h1,h1): Sweden. Torne Lappmark, Jukkasjärvi, Vastakielisenvaara; 2002, LH; S: B73007; AY625998, MZ447579. M858 (A:h6,h2): Sweden. Lycksele lappmark, Tärna, Atofjället; 2012, LH et al.; S: B195253; ON323950, ON340919. M1046 (B:h2,h2): Sweden. Hälsingland, Los, Fräkentjärn; 2008, LH et al.; S: B139054; ON323951, ON340920. M1383 (A:h3,h1): Svalbard. Billefjorden, Adolfbukta, Thomsonelva; 2019, LH & I.Bisang; S: B290588; ON323952, ON340921. M1385 (D:h1,h2): Sweden. Södermanland, Åker, Strålsjömossen; 2019, LH; S: B290657; ON323953, ON340922. M1386 (D:h1,h2): Sweden. Dalarna, Boda, Ensänget; 2018, LH; S: B284135; ON323954, ON340923. M1387 (D:h1,h2): Sweden. Härjedalen, Sveg, Duvberg; 2018, LH et al.; S: B289222; ON323955, ON340924. M1388 (B:h2,h2): Sweden. Torne lappmark, Karesuando, Tsuonamavaara; 2019, LH et al.; S: B291153; ON323956, ON340925. M1389 (A:h3,h3): Svalbard. Longyearbyen, Longyearbyen; 2019, LH & I.Bisang; S: B290578; ON323957, ON340926. M1392 (A:h3,h2): Svalbard. Longyearbyen, Björndalen; 2019, LH & I.Bisang; S: B290613; ON323958, ON340927. M1433 (B:h2,h2): Svalbard. Kap Waern; 1958, O.Rønning; TRH: 74807; ON323959, ON340928. M1593 (B:h2,h2): Norway. Vågå, Maurvangen; 2016, T.Peterka et al.; S: B311335; ON323961, ON340930. M1594 (D:h1,h2): Finland. Enontekiö, Leppäjärvi; 2019, Z.Plesková et al.; S: B311338; ON323962, ON340931. M1595 (C:h4,h2): Sweden. Härjedalen, Lillhärdal, Härjån-Bäreggen; 2018, LH et al.; S: B289058; ON323963, ON340932. M1596 (B:h2,h2): Sweden. Jämtland, Åre, Storvallen; 2020, LH & I.Bisang; S: B299271; ON323964, ON340933. M1597 (C:h4,h4): Sweden. Jämtland, Åre, Rundvalen; 2020, LH & I.Bisang; S: B299274; ON323965, ON340934. M1598 (A:h3,h2): Sweden. Jämtland, Undersåker, Långtjärnen; 2020, LH & I.Bisang; S: B299282; ON323966, ON340935. M1599 (D:h1,h2): Sweden. Medelpad, Borgsjö, Gammelboda; 2019, LH; S: B292051; ON323967, ON340936. M1600 (D:h1,h2): Sweden. Medelpad, Borgsjö, Mallberget; 2019, LH; S: B292069; ON323968, ON340937. M1601 (C:h4,h4): Sweden. Medelpad, Borgsjö, Slåttmyran; 2019, LH; S: B292139; ON323969, ON340938. M1602 (B:h2,h2): Sweden. Torne lappmark, Karesuando, Árbuvuopmi; 2019, LH et al.; S: B291084; ON323970, ON340939. M1603 (B:h2,h2): Sweden. Torne lappmark, Karesuando, Árbujávri; 2019, LH et al.; S: B291093; ON323971, ON340940. M1604 (B:h2,h3): Sweden. Torne lappmark, Karesuando, Maunu; 2019, LH et al.; S: B291102; ON323972, ON340941. M1605 (B:h2,h2): Sweden. Torne lappmark, Karesuando, Ainattivaara; 2019, LH et al.; S: B291111; ON323973, ON340942. M1606 (A:h3,h2): Sweden. Torne lappmark, Karesuando, Buolžajávri; 2019, LH et al.; S: B291135; ON323974, ON340943. M1607 (B:h2,h2): Sweden. Torne lappmark, Karesuando, Siilaslompolo; 2019, LH et al.; S: B291164; ON323975, ON340944. M1608 (D:h1,h2): Sweden. Värmland, Töcksmark, Tutjärnen; 2012, N.Lönnell; S: B299257; ON323976, ON340945. M1609 (D:h1,h2): Sweden. Värmland, Södra Finnskoga, Bredsjön; 2012, N.Lönnell; S: B299258; ON323977, ON340946. M1610 (A:h5,h2): Sweden. Värmland, Norra Ny, Stavsjön; 2012, N.Lönnell; S: B299259; ON323978, ON340947. M1611 (A:h5,h2): Sweden. Medelpad, Ljustorp, Storfuskberg; 2012, N.Lönnell; S: B299260; ON323979, ON340948. M1612 (D:h1,h2): Sweden. Medelpad, Ljustorp, Ålidberget; 2012, N.Lönnell; S: B299261; ON323980, ON340949. M1613 (D:h1,h2): Sweden. Hälsingland, Enånger, Yxberg; 2012, N.Lönnell; S: B299262; ON323981, ON340950. M1614 (A:h5,h2): Sweden. Hälsingland, Delsbo, Prästvallen; 2012, N.Lönnell; S: B299263; ON323982, ON340951. M1615 (A:h5,h2): Sweden. Ångermanland, Viksjö, Amerika; 2012, N.Lönnell; S: B299264; ON323983, ON340952. M1616 (B:h2,h2): Sweden. Västerbotten, Byske, Sörviksmyren; 2012, N.Lönnell; S: B299265; ON323984, ON340953. M1617 (B:h2,h2): Sweden. Västerbotten, Burträsk, Brännvinshålet; 2012, N.Lönnell; S: B299266; ON323985, ON340954. M1618 (B:h2,h2): Sweden. Dalarna, Äppelbo, Gröntjärnen; 2012, N.Lönnell; S: B299267; ON323986, ON340955. M1619 (D:h1,h2): Sweden. Härjedalen, Älvros, Sätermyren; 2012, N.Lönnell; S: B299268; ON323987, ON340956. M1620 (A:h3,h2): Sweden. Ångermanland, Bjurholm, Brånamyran; 2012, N.Lönnell; S: B299269; ON323988, ON340957. M1621 (D:h1,h2): Sweden. Jämtland, Oviken, Fittjebodarna; 2012, N.Lönnell; S: B299270; ON323989, ON340958. M1622 (D:h1,h2): Sweden. Västmanland, Västanfors, Kolarbysjön; 2020, LH; S: B299469; ON323990, ON340959. M1623 (D:h1,h2): Sweden. Västmanland, Norberg, Tattarberget; 2020, LH; S: B299473; ON323991, ON340960. M1625 (A:h5,h2): Sweden. Västmanland, Norberg, St. Malmkärra; 2020, LH; S: B299476; ON323992, ON340961. M1666 (D:h1,h2): Sweden. Jämtland, Kall, Juvuln; 2020, LH et al.; S: B299892; ON323993, ON340962. M1667 (B:h2,h2): Sweden. Jämtland, Kall, Torrön; 2020, LH et al.; S: B299893; ON323994, ON340963. M1668 (B:h2,h2): Sweden. Jämtland, Kall, Torrön; 2020, LH et al.; S: B299898; ON323995, ON340964. M1669 (B:h2,h2): Sweden. Jämtland, Kall, Torrön; 2020, LH et al.; S: B299905; ON323996, ON340965. M1670 (D:h1,h2): Sweden. Jämtland, Kall, Långtjärnen; 2020, LH et al.; S: B299917; ON323997, ON340966. M1671 (C:h4,h2): Sweden. Jämtland, Kall, Åbo; 2020, LH et al.; S: B299918; ON323998, ON340967. M1672 (C:h4,h4): Sweden. Jämtland, Kall, Strytjärnen-Rutsvallen; 2020, LH et al.; S: B299921; ON323999, ON340968. M1673 (D:h1,h2): Sweden. Jämtland, Kall, Jävsjön; 2020, LH et al.; S: B299926; ON324000, ON340969. M1674 (D:h1,h2): Sweden. Jämtland, Kall, Juvuln; 2020, LH et al.; S: B299936; ON324001, ON340970. M1675 (D:h1,h2): Sweden. Jämtland, Kall, Movallen-Bergsjön; 2020, LH et al.; S: B299947; ON324002, ON340971. M1699 (D:h1,h2): Sweden. Västmanland, Hällefors, Jungfrutjärnarna; 2020, LH; S: B301004; ON324003, ON340972. M1700 (D:h1,h2): Sweden. Västmanland, Hjulsjö, Björksjön; 2020, LH; S: B301006; ON324004, ON340973. M1701 (C:h4,h2): Sweden. Västmanland, Hjulsjö, Björksjön; 2020, LH; S: B301008; ON324005, ON340974. M1702 (D:h1,h2): Sweden. Västmanland, Hällefors, Södra Masbo; 2020, LH; S: B301010; ON324006, ON340975. M1703 (D:h1,h2): Sweden. Värmland, Färnebo, Östra Björntjärnen; 2020, LH; S: B301015; ON324007, ON340976. M1704 (D:h1,h2): Sweden. Värmland, Färnebo, Hungern; 2020, LH; S: B301036; ON324008, ON340977. M1709 (B:h2,h2): Sweden. Pite lappmark, Arvidsjaur, 550 m SW Pite älv; 2021, LH & A.Jörgensen; S: B305340; ON324009, ON340978. M1710 (B:h2,h2): Sweden. Lule lappmark, Jokkmokk, Karats; 2021, LH & A.Jörgensen; S: B305348; ON324010, ON340979. M1711 (C:h4,h4): Sweden. Lule lappmark, Jokkmokk, Karats; 2021, LH & A.Jörgensen; S: B305355; ON324011, ON340980. M1712 (D:h1,h2): Sweden. Norrbotten, Edefors, Övre Svartlå; 2021, LH & A.Jörgensen; S: B305357; ON324012, ON340981. M1713 (B:h2,h2): Sweden. Lule lappmark, Jokkmokk, Goabddális; 2021, LH & A.Jörgensen; S: B305433; ON324013, ON340982. OUTGROUP: Scorpidium cossonii (Schimp.) Hedenäs: Sweden. Torne Lappmark, Jukkasjärvi, Torneträsk area, Kaisepakte; 2017, LH; S: B254928; MK456245, MK466720.
Appendix 2. Specimen data and GenBank accession numbers for specimens of Scorpidium cossonii and Sarmentypnum exannulatum from the northern Scandinavian lowland (NLO), southern Scandinavian mountains (SMO) and southern Scandinavian lowland (SLO) regions. Data format: Specimen no. Locality (all in Sweden); Collection year, Collector (collector’s no. [LH = L.Hedenäs]); S herbarium registration no.; GenBank accession nos for ITS, rpl16 (Sc. cossonii) or ITS, rpl16, trnG and trnL–trnF (Sa. exannulatum).
Scorpidium cossonii (Schimp.) Hedenäs. NLO: M3. Norrbotten, Tärendö; 1990, LH & M.Aronsson NT90-795; B41603; DQ393039, DQ405089. M4. Norrbotten, Pajala; 1990, LH & M.Aronsson NT90-196; B41600; DQ393040, DQ405090. M5. Norrbotten, Pajala; 1990, LH & M.Aronsson NT90-89; B41601; DQ393041, DQ405091. M276. Torne lappmark, Jukkasjärvi; 2002, LH; B73001; DQ393111, DQ405162. M277. Torne lappmark, Jukkasjärvi; 2002, LH; B73002; DQ393112, DQ405163. M278. Torne lappmark, Jukkasjärvi; 2002, LH; B73003; DQ393113, DQ405164. M279. Torne lappmark, Jukkasjärvi; 2002, LH; B73004; DQ393114, DQ405165. M280. Torne lappmark, Jukkasjärvi; 2002, LH; B73005; DQ393115, DQ405166. M860. Lycksele lappmark, Malå; 2016, LH & G.Odelvik; B237109; MK456223, MK466698. M862. Lycksele lappmark, Lycksele; 2016, LH & G.Odelvik; B242117; MK456225, MK466700. M863. Norrbotten, Mäntyvaraanvuoma; 1998, O.Johansson; B134106; MK456226, MK466701. M868. Västerbotten, Norsjö; 2016, LH& G.Odelvik; B236867; MK456231, MK466706. M939. Lule lappmark, Lautavouma; 1996, S.Backe; B134181; MK456233, MK466708. M940. Lule lappmark, Kuosatjape; 1999,O.Johansson; B134182; MK456234, MK466709. M942. Lule lappmark, Kurtakkovuoma; 2001, S.Thuresson; B134615; MK456235, MK466710. M943. Lule lappmark, Jokkmokk; 2017, LH; B253137; MK456236, MK466711. M944. Lule lappmark, Jokkmokk; 2017, LH; B253145; MK456237, MK466712. M945. Lule lappmark, Gällivare; 2017, LH; B253202; MK456238, MK466713. M946. Lule lappmark, Gällivare; 2017, LH; B253211; MK456239, MK466714. M947. Lule lappmark, Gällivare; 2017, LH; B253213; MK456240, MK466715. M948. Torne lappmark, Paanavuoma; 2000, VMI Norrbotten; B134717; MK456241, MK466716. M1039. Lycksele lappmark, Lycksele; 2018, LH; B279841; MK456255, MK466730. M1040. Lycksele lappmark, Lycksele; 2018, LH; B279853; MK456256, MK466731. SMO: M7. Jämtland, Frostviken; 1997, LH & A.Kooijman; B1434; DQ393043, DQ405092. M60. Jämtland, Frostviken; 1997, LH & A.Kooijman; B1469; DQ393051, DQ405101. M108. Jämtland, Åre; 2001, LH; B61795; DQ393065, DQ405116. M109. Jämtland, Åre; 2001, LH; B61796; DQ393066, DQ405117. M113. Jämtland, Åre; 2001, LH; B61800; DQ393067, DQ405118. M115. Jämtland, Åre; 2001, LH; B61803; DQ393068, DQ405119. M117. Jämtland, Åre; 2001, LH; B61805; DQ393069, DQ405120. M119. Jämtland, Åre; 2001, LH; B61807; AY625996, DQ304458. M123. Jämtland, Åre; 2001, LH; B61813; DQ393070, DQ405121. M130. Jämtland, Undersåker; 2001, LH; B61821; DQ393071, DQ405122. M132. Jämtland, Undersåker; 2001, LH; B61829; DQ393072, DQ405123. M134. Jämtland, Undersåker; 2001, LH; B61831; DQ393073, DQ405124. M136. Jämtland, Undersåker; 2001, LH; B61833; DQ393074, DQ405125. M140. Jämtland, Undersåker; 2001, LH; B61837; DQ393075, DQ405126. M143. Jämtland, Undersåker; 2001, LH; B61840; DQ393076, DQ405127. M146. Jämtland, Mörsil; 2001, LH; B61843; DQ393077, DQ405128. M152. Jämtland, Mörsil; 2001, LH; B61849; DQ393078, DQ405129. M163. Jämtland, Mattmar; 2001, LH; B61861; DQ393079, DQ405130. M853. Härjedalen, Tännäs; 2014, LH; B207525; MK456217, MK466692. M854. Härjedalen, Tännäs; 2014, LH; B207597; MK456218, MK466693. M869. Åsele lappmark, Vilhelmina; 2004, LH; B95099; MK456232, MK466707. M1047. Jämtland, Frostviken; 2009, LH; B164614; MK456262, MK466737. SLO: M63. Västergötland, Ottravad; 1996, LH & A.Kooijman; B1074; DQ393054, DQ405104. M99. Uppland, Djurö; 2001, LH; B62286; DQ393061, DQ405112. M103. Uppland, Djurö; 2001, LH; B62287; DQ393062, DQ405113. M104. Uppland, Djurö; 2001, LH; B62288; DQ393063, DQ405114. M106. Uppland, Djurö; 2001, LH; B62289; DQ393064, DQ405115. M169. Västergötland, Skövde; 2001, LH; B61976; DQ393080, DQ405131. M173. Västergötland, Varnhem; 2001, LH; B61983; DQ393081, DQ405132. M174. Västergötland, Norra Lundby; 2001, LH; B61985; DQ393082, DQ405133. M179. Södermanland, Hölö; 2001, LH; B62801; DQ393083, DQ405134. M209. Uppland, Börstil; 2002, LH; B70219; DQ393089, DQ405140. M212. Uppland, Hökhuvud; 2002, LH; B70222; DQ393090, DQ405141. M1050. Västmanland, Nora; 2015, LH; B226643; MK456265, MK466740.
Sarmentypnum exannulatum (Schimp.) Hedenäs. NLO: M71. Åsele lappmark, Vilhelmina; 1991, LH & A.Kooijman; B21481; DQ400008, DQ405180, DQ404995, DQ404939. M75. Åsele lappmark, Vilhelmina; 1991, LH; B58535; DQ400011, DQ405182, DQ404999, DQ404943. M78. Lule lappmark, Vuollerim; 1991, LH; B58538; DQ400014, DQ405183, DQ405002, DQ404946. M319. Torne lappmark, Jukkasjärvi; 2002, LH; B73044; DQ400043, DQ405212, DQ405031, DQ404975. M320. Torne lappmark, Jukkasjärvi; 2002, LH; B73045; DQ400044, DQ405213, DQ405032, DQ404976. M322. Torne lappmark, Jukkasjärvi; 2002, LH; B73047; DQ400046, DQ405215, DQ405034, DQ404978. M323. Torne lappmark, Jukkasjärvi; 2002, LH; B73048; DQ400047, DQ405216, DQ405035, DQ404979. M324. Torne lappmark, Jukkasjärvi; 2002, LH; B73049; DQ400048, DQ405217, DQ405036, DQ404980. M325. Torne lappmark, Jukkasjärvi; 2002, LH; B73050; DQ400049, DQ405218, DQ405037, DQ404981. M326. Torne lappmark, Jukkasjärvi; 2002, LH; B73051; DQ400050, DQ405219, DQ405038, DQ404982. M327. Torne lappmark, Jukkasjärvi; 2002, LH; B73052; DQ400051, DQ405220, DQ405039, DQ404983. M328. Torne lappmark, Jukkasjärvi; 2002, LH; B73053; DQ400052, DQ405221, DQ405040, DQ404984. M329. Torne lappmark, Jukkasjärvi; 2002, LH; B73054; DQ400053, DQ405222, DQ405041, DQ404985. M330. Torne lappmark, Jukkasjärvi; 2002, LH; B73055; DQ400054, DQ405223, DQ405042, DQ404986. M331. Torne lappmark, Jukkasjärvi; 2002, LH; B73056; DQ400055, DQ405224, DQ405043, DQ404987. M332. Torne lappmark, Jukkasjärvi; 2002, LH; B73057; DQ400056, DQ405225, DQ405044, DQ404988. M333. Torne lappmark, Jukkasjärvi; 2002, LH; B73058; DQ400057, DQ405226, DQ405045, DQ404989. M875. Lule lappmark, Palagekyrkan; 1996, M.Norin; B117722; MK456177, MK466652, MK467078, MK467171. M879. Lycksele lappmark, Malå; 2016, LH & G.Odelvik; B240965; MK456181, MK466656, MK467082, MK467175. M880. Norrbotten, Tervajänkkä; 1995, S.Backe; B117754; MK456182, MK466657, MK467083, MK467176. M885. Torne lappmark, Karesuando; 1990, LH & M.Aronsson; B42429; MK456187, MK466662, MK467088, MK467181. M886. Torne lappmark, Karesuando; 1990, LH & M.Aronsson; B42431; MK456188, MK466663, MK467089, MK467182. M887. Torne lappmark, Karesuando; 1990, LH & M.Aronsson; B42432; MK456189, MK466664, MK467090, MK467183. M888. Torne lappmark, Karesuando; 1990, LH & M.Aronsson; B242902; MK456190, MK466665, MK467091, MK467184. M889. Torne lappmark, Karesuando; 1990, LH & M.Aronsson; B242904; MK456191, MK466666, MK467092, MK467185. M890. Västerbotten, Norsjö; 2016, LH & G.Odelvik; B240430; MK456192, MK466667, MK467093, MK467186. M929. Lule lappmark, Jokkmokk; 2017, LH; B253151; MK456194, MK466669, MK467095, MK467188. M930. Lule lappmark, Jokkmokk; 2017, LH; B253159; MK456195, MK466670, MK467096, MK467189. M931. Lule lappmark, Jokkmokk; 2017, LH; B253162; MK456196, MK466671, MK467097, MK467190. M932. Lule lappmark, Jokkmokk; 2017, LH; B253167; MK456197, MK466672, MK467098, MK467191. M933. Lule lappmark, Gällivare; 2017, LH; B253180; MK456198, MK466673, MK467099, MK467192. M934. Lule lappmark, Gällivare; 2017, LH; B253184; MK456199, MK466674, MK467100, MK467193. M935. Lule lappmark, Gällivare; 2017, LH; B253191; MK456200, MK466675, MK467101, MK467194. M936. Lule lappmark, Gällivare; 2017, LH; B253214; MK456201, MK466676, MK467102, MK467195. M937. Lule lappmark, Gällivare; 2017, LH; B253219; MK456202, MK466677, MK467103, MK467196. M938. Lule lappmark, Gällivare; 2017, LH; B253223; MK456203, MK466678, MK467104, MK467197. M1036. Lycksele lappmark, Lycksele; 2018, LH; B279846; MK456213, MK466688, MK467114, MK467207. SMO: M112. Jämtland, Åre; 2001, LH; B61799; DQ400017, DQ405186, DQ405005, DQ404949. M120. Jämtland, Åre; 2001, LH; B61808; DQ400018, DQ405187, DQ405006, DQ404950. M124. Jämtland, Åre; 2001, LH; B61815; DQ400019, DQ405188, DQ405007, DQ404951. M126. Jämtland, Åre; 2001, LH; B61817; DQ400020, DQ405189, DQ405008, DQ404952. M128. Jämtland, Undersåker; 2001, LH; B61819; DQ400021, DQ405190, DQ405009, DQ404953. M129. Jämtland, Undersåker; 2001, LH; B61820; DQ400022, DQ405191, DQ405010, DQ404954. M131. Jämtland, Undersåker; 2001, LH; B61828; DQ400023, DQ405192, DQ405011, DQ404955. M144. Jämtland, Undersåker; 2001, LH; B61841; DQ400024, DQ405193, DQ405012, DQ404956. M156. Jämtland, Mörsil; 2001, LH; B61854; DQ400025, DQ405194, DQ405013, DQ404957. M160. Jämtland, Mörsil; 2001, LH; B61858; DQ400026, DQ405195, DQ405014, DQ404958. M161. Jämtland, Mörsil; 2001, LH; B61859; DQ400027, DQ405196, DQ405015, DQ404959. M164. Jämtland, Mattmar; 2001, LH; B61862; DQ400028, DQ405197, DQ405016, DQ404960. M870. Härjedalen, Tännäs; 2005, LH; B104251; MK456172, MK466647, MK467073, MK467166. M872. Jämtland, Åre; 2010, LH; B177209; MK456174, MK466649, MK467075, MK467168. SLO: M72. Västergötland, Gustav Adolf; 1993, LH & A.Kooijman; B58530; DQ400009, DQ405181, DQ404996, DQ404940. M100. Uppland, Djurö; 2001, LH; B62290; DQ400015, DQ405184, DQ405003, DQ404947. M102. Uppland, Djurö; 2001, LH; B62291; DQ400016, DQ405185, DQ405004, DQ404948. M170. Västergötland, Skövde; 2001, LH; B61978; DQ400029, DQ405198, DQ405017, DQ404961. M172. Västergötland, Stenstorp; 2001, LH; B61982; DQ400030, DQ405199, DQ405018, DQ404962. M176. Västergötland, Norra Lundby; 2001, LH; B61988; DQ400031, DQ405200, DQ405019, DQ404963. M177. Västergötland, Norra Lundby; 2001, LH; B61989; AY625997, DQ307051, DQ304462, AY626015. M178. Västergötland, Segerstad; 2001, LH; B61991; DQ400032, DQ405201, DQ405020, DQ404964. M339. Södermanland, Nacka; 2002, LH; B73427; DQ400062, DQ405231, DQ405050, DQ404994. M395. Uppland, Singö; 2000, LH; B32426; FJ474535, FJ474662, FJ474786, FJ474409. M396. Uppland, Jumkil; 1987, LH; B87658; FJ474536, FJ474663, FJ474787, FJ474410. M397. Östergötland, Krokek; 1998, LH; B13944; FJ474537, FJ474664, FJ474788, FJ474411. M541. Västergötland, Brandstorp; 2004, LH; B91693; FJ474590, FJ474714, FJ474840, FJ474463. M542. Västergötland, Brandstorp; 2004, LH; B91694; FJ474591, FJ474715, FJ474841, FJ474464. M544. Västergötland, Habo; 2004, LH; B91695; FJ474592, FJ474716, FJ474842, FJ474465. M545. Västergötland, Habo; 2004, LH; B91695; FJ474593, FJ474717, FJ474843, FJ474466. M546. Västergötland, Habo; 2004, LH; B91696; FJ474594, FJ474718, FJ474844, FJ474467. M547. Västergötland, Habo; 2004, LH; B91697; FJ474595, FJ474719, FJ474845, FJ474468. M548. Småland, Norra Unnaryd; 2004, LH; B91698; FJ474596, FJ474720, FJ474846, FJ474469. M549. Småland, Norra Unnaryd; 2004, LH; B91699; FJ474597, FJ474721, FJ474847, FJ474470.
