Abstract
In this study, reverse transcription PCR (RT–PCR) detection revealed a high infection rate of Grapevine virus A (GVA) in grapevine plants cultivated in China. Population structures of 14 GVA isolates from Chinese grapevines were studied by single-stranded conformation polymorphism (SSCP) of the coat protein (CP) gene. Results showed that while most isolates contained a population of sequence variants, with one being predominant, one isolate (AF) had three major variants, and two isolates (BSSL and BSBD) consisted of a number of sequence variants with almost the same detection frequencies. The estimation of genetic diversity intra isolates by sequence alignment analysis indicated a high variability in the CP gene and the occurrence of mixed infections in some grapevines by divergent sequence variants. RT–PCR analysis using variant-specific primers (H587/C995, GVA6591F/GVA6906R) and phylogenetic analyses of CP gene sequences suggested that all sequence variants from Chinese isolates were separated into two subgroups IA and IB in the group I.
Résumé
Cette étude, faisant appel à la technique de RT–PCR, a révélé un taux élevé d'infection causée par le virus A de la vigne (GVA) dans les vignes cultivées en Chine. Les structures de populations de 14 isolats de GVA issus de vignes chinoises ont été étudiées en fonction du polymorphisme de conformation des ADN simples brins du gène de la protéine capsidique (PC). Les résultats ont montré que, même si la plupart des isolats contenaient une population de variants de séquence, dont un prédominant, un isolat (AF) comprenait trois variants majeurs et deux isolats (BSSL et BSBD) comptaient un nombre de variants de séquence affichant sensiblement les mêmes fréquences de détection. L’évaluation de la diversité génétique chez les isolats, effectuée par l'analyse de l'alignement des séquences, a affiché un haut degré de variabilité chez le gène PC et démontré l'incidence d'infection mixte causée par des variants de séquence divergents chez certaines vignes. L'analyse par RT–PCR avec amorces spécifiques des variants (H587/C995, GVA6591F/GVA6906R) et les analyses phylogénétiques des séquences du gène PC ont semblé indiquer que tous les variants de séquence issus des isolats chinois étaient divisés en deux sous-groupes, IA et IB, du groupe I.
Introduction
Rugose wood complex (RW) is among the most widespread and devastating disease syndromes affecting grapevines (Martelli, Citation1993). Grapevine virus A (GVA) is closely associated with the development of RW disorders (Garau et al., Citation1994; Chevalier et al., Citation1995; Choueiri et al., Citation1997). GVA, the type member of genus Vitivirus in the family Betaflexiviridae (Martelli et al., Citation1997;
Carstens, Citation2010), is the most prevalent virus of grapevine and is commonly detected in cultivated grapevines. The virus has filamentous particles about 800 nm long. To date, genomes of 11 GVA isolates have been completely sequenced (Minafra et al., Citation1997; Galiakparov et al., Citation2003; Goszczynski et al., Citation2008). The genome of GVA consists of positive single-stranded RNA, and is organized into five open reading frames (ORF) (Minafra et al., Citation1997), which encode the putative replicase (ORF1), a polypeptide (ORF2) whose function is unknown, the putative movement protein (MP) (ORF3), the coat protein (CP) (ORF4) and the putative nucleic acid-binding protein (ORF5), respectively. The virus is transmissible to the herbaceous host Nicotiana benthamiana L. by insect vectors (Rosciglione & Castellano, Citation1985) or by mechanical inoculation (Monette & James, Citation1990) and disseminated by infected propagating materials. The reported vectors include mealybugs Pseudococcid Pseudococcus longispinus, Ps. affinis, Planococcus ficus and Heliococcus bohemicus (Rosciglione et al., Citation1983; Garau et al., Citation1995; La Notte et al., Citation1997; Zorloni et al., Citation2006), soft scale mealybugs Neopulvinaria innumertabitis (Fortusini et al., Citation1997) and larvae of Parthenolecanium corni (Hommay et al., Citation2008).
Recent studies revealed that the virus is highly heterogenic, and divergent variants were identified from different countries (Monette & James, Citation1990; Goszczynski & Jooste, Citation2003 a, Citation2003 b; Murolo et al., Citation2008; Goszczynski et al., Citation2008). Analysis based on overall nucleotide (nt) sequence identity in the 3′ terminal part of the GVA genome revealed that GVA isolates clustered into three molecular groups (I, II, III), sharing 91.0–99.8% sequence identity within groups and 78.0–89.3% sequence identity between groups. Group III consists of mild isolates, which shared 97.7–99.8% within group and only 78.0–79.6% sequence identity between groups (Goszczynski & Jooste, 2003a). Goszczynski (Citation2007) reported that GVA variants of group II are closely associated with Shiraz disease (SD) occurring in South Africa.
The RT–PCR technique, based on universal primers designed by MacKenzie (Citation1997), has been widely used to detect all identified variants of GVA. Primer sets for the detection of variants of group I and II (Minafra & Hadidi, Citation1994) or group III (Goszczynski & Jooste, 2003a), respectively, were also developed and applied for the analysis of GVA variants. Single-stranded conformation polymorphism (SSCP) is a useful tool for the identification of different virus variants (Magome et al., Citation1999; Rubio et al., 2001; Goszczynski & Jooste, 2003b; Kong et al., Citation2005; Turturo et al., Citation2005; Ayllón et al., Citation2006; Goszczynski, Citation2007). RT–PCR combined with SSCP analysis for GVA isolates recovered from N. benthamiana revealed extensive molecular heterogeneity of the virus (Goszczynski & Jooste, 2003a, 2003b). Furthermore, it was observed that the SSCP patterns of a 234-nt fragment from the ORF3 were indicative of GVA variants of molecular group II (Goszczynski, Citation2007). Recently, RT–PCR–RFLP analysis of the coat protein (CP) gene has also been used for the characterization of the genetic and population diversity of GVA. Murolo et al. (Citation2008) divided group I into two subgroups and reported a new putative group IV which showed RFLP pattern E.
Grapevine has been widely grown in China for a long time. It has been noticed that RW occurs in grafted grapevine cultivars and severely affects the growth of grapevine plants (Wang et al., 1996). For self-rooting plants, which usually are symptomless, the dissemination of virus-infected material may result in occurrence of the disease in new areas. GVA has been detected from cultivated grapevine plants in different regions in China. However, only partial molecular information is known on its CP gene (Wang et al., 2008). In this paper, the population structure and genetic variability of GVA isolates from cultivated grapevine plants grown in three Chinese provinces were analyzed by clone-based SSCP and sequence analyses of the coat protein gene. The results give a general view of the genetic diversity of GVA isolates from China and provide useful information for effective detection and for preventing the dissemination of the viral disease in China.
Materials and methods
Virus sources
Dormant canes of 55 grapevine varieties were collected randomly from three vineyards in China, 32 from Zhengzhou in Henan province, four from Xingcheng in Liaoning province and 19 from Wuhan in Hubei province. All grapevines collected displayed no visible symptoms in these vineyards.
DsRNA extraction
Approximately 1 g cortical scrapings from dormant canes were homogenized in liquid nitrogen, and dsRNA was obtained by chloroform extraction and purification through cellulose CF-11 (Whatman) as described by Rezaian et al. (Citation1991).
RT–PCR and cloning
Approximately 600 ng dsRNA, 1 μL 6-mer random primers (TaKaRa, Dalian, China) and RNase-free water were mixed to a final volume of 10 μL. The mixture was denatured at 99 °C for 5 min, and chilled on ice. cDNA synthesis was performed with M-MLV reverse transcriptase according to the instructions of the manufacturer (Promega, Madison, USA).
Universal primer set H7038/C7273 (MacKenzie, Citation1997) was used for detection of GVA. Primer sets H587/C995 (Minafra & Hadidi, Citation1994) and 6591F/6906R (Goszczynski & Jooste, 2003a) were used for the grouping of GVA isolates. The primer set ACL/ACR (5′-GCTGGG GTTGAAGACAAATG-3′/5′-GCGAGAAACGATGGGTCAT-3′) for amplification of complete CP gene was designed based on the conserved region flanking the CP gene (GenBank accession no. DQ855081, DQ855085, AY244516, AF007415 and NC_003604).
PCR reactions were taken place in a 20 μL volume with reaction mixtures containing 2 μL of 10 × PCR buffer, 0.2 mM of dNTPs, 0.2 mM of each primer, one unit of Taq DNA polymerase (TaKaRa, Dalian, China), and 3 μL cDNA. Reactions were conducted in a PCR Thermal Cycler (Model PTC-200, MJ Research, USA). The PCR reaction condition was as follows: 94 °C for 5 min, followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 45 s at 53 °C for primer sets H7038/C7273 and 6591F/6906R or at 56 °C for primer sets H587/C995 and ACL/ACR, extension for 1 min at 72 °C, and final extension for 10 min at 72 °C. The PCR products were separated by electrophoresis in 1.2% agarose gel, stained with ethidium bromide and visualized under UV light.
Amplicons of GVA CP genes were gel purified and inserted into the PMD18-T vector (Takara, Dalian, China) following the manufacturer's instructions. The recombinant plasmids were transformed into Escherichia coli DH5α.
SSCP analysis and sequencing
Six to 23 positive clones of RT-PCR products of GVA CP gene from each isolate were selected randomly for SSCP analysis according to the method of Palacio & Duran-Vila (Citation1999). The cloned CP gene was amplified from each of selected positive clones with primer set ACL/ACR. An aliquot of 1 μL PCR product was mixed with 2 μL denaturing solution [95% deionized formamide, 10 mM NaOH, 0.05% bromophenol blue] boiled for 10 min, chilled on ice immediately for 2 min and separated by 8% non-denaturing polyacrylamide gel electrophoresis at 200V at 4 °C for 18 h. SSCP profiles were visualized by silver staining. The PCR product showing unique SSCP band pattern was designated a haplotype. Clones representing different SSCP haplotypes were sent to Genscript Corporation (Nanjing, China) for sequencing.
Nucleotide sequences analyses
Multiple nucleotide alignments were conducted using CLUSTALX 1.8 (Thompson et al., Citation1997). Phylogenetic tree was constructed with neighbour-joining method using MEGA 4.1 with 1000 bootstrap replicates to assess the robustness of the nodes (Tamura et al., Citation2007). Nucleotide distance with stand error was evaluated according to the method of Juke & Cantor (Citation1969) for correction of superimposed substitutions with MEGA 4.1.
Results and discussion
RT–PCR detection of GVA isolates and molecular grouping of GVA isolates
Forty-four out of 55 grapevine samples tested positive for GVA as detected by RT–PCR with the set H7038/C7273, a universal primer set for GVA (MacKenzie, Citation1997). It was noticed that grapevine plants from a germplasm collection in Henan province had a high GVA infection rate of 90.6%. The high infection frequency might be associated with the wide occurrence of mealybugs in the vineyard (unpublished observations). Rugose wood (RW) disorders which are closely associated with GVA infection (Garau et al., Citation1994) had been previously recorded for graft-propagated grapevine plants from Liaoning and Shandong provinces in China (Wang et al., Citation1996). GVA was also detected in Sinkiang and Sichuan province (Ribeiro et al., Citation2004; Wang et al., Citation2008). Previous reports suggested a wide distribution of GVA among grapevines grown in China.
Further grouping analysis with two group-specific primer sets showed that H587/C995 amplified 100% of these isolates, indicating that the GVA isolates from these 44 grapevine samples belong to group I or II. On the other hand, the primer set 6591F/6906R did not yield any amplification from these samples, indicating the absence of group III GVA variants (data not shown).
Intra-isolate population structure of GVA revealed by SSCP and genetic distance analysis
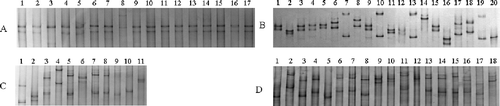
The 637 bp DNA products containing complete CP gene (597 bp) amplified from 14 GVA isolates were cloned. A total of 254 positive clones (6 to 23 clones for each isolate) were randomly selected for primary intra-isolate population structure analysis by SSCP. Results showed that these isolates had highly divergent population structures. Two categories were observed. In isolates AF, BSBD, BSSL, DB, FB, GFMG and SFLS, the ratio of SSCP haplotypes/tested clones was over 50% and in the remaining seven isolates the ratio was below 40%. Eleven isolates had one predominant haplotype, and the highest frequency of predominant variant appeared in isolate HBS () (88.2 % of 17 tested clones) (). Nevertheless, the isolate AF () contained three major haplotypes, accounting for 4, 3 and 3 out of 20 tested clones, respectively. The population structures of isolates BSSL and BSBD ( and d) were even more complex, each clone represented a haplotype in BSSL isolate, and 16 SSCP haplotypes out of 18 clones of BSBD were detected ().
Table 1. Analyses of SSCP profiles of cloned RT-PCR products of CP genes, and genetic diversity among CP haplotypes from each of 14 GVA isolates
Fig. 1. SSCP analyses for coat protein genes cloned from four Grapevine virus A isolates showing different population structures. Numbers above each lane refer to different clones. a and b, show simple SSCP types with predominant haplotypes from isolates HBS and AF. c and d, show complex SSCP types without predominant haplotypes from isolates BSSL and BSBD.
A total of 114 clones with different SSCP patterns from each isolate were sequenced and genetic diversities intra-isolates were evaluated. Results showed that seven out of 14 isolates contained genetically similar sequence variants with maximum values of intra-isolate diversity ranging from 0.0095 ± 0.0036 to 0.0554 ± 0.0092 (). In isolate BSSL, although each of 11 selected clones showed different SSCP patterns, the genetic diversity among them was very low, ranging from 0.0032 ± 0.0022 to 0.050 ± 0.0090. Similarly, isolates DBPT, HBS, HU, HZJM, MFLT and SFLS had low inter-clone diversities, which were less than 0.0554. In contrast, clones obtained from seven other isolates were heterogeneous and had significantly higher inter-clone genetic diversities, ranging from 0.0780 ± 0.0121 to 0.1693 ± 0.0202 (). These seven isolates were considered to have two sub-isolates. Genetic diversities between sub-isolates were higher than 0.0780 ± 0.0121. These results indicated the occurrence of mixed infection of highly divergent GVA variants in some plants. Mixed infection is prevalent in viruses which infect perennial plants. It might be due to the reduplicative inoculation by insect vectors (Ayllón et al., Citation2001; Angelini et al., Citation2004; Vigne et al., Citation2004; Fattouch et al., Citation2005).
Phylogenetic analysis of representative molecular variants from different GVA isolates
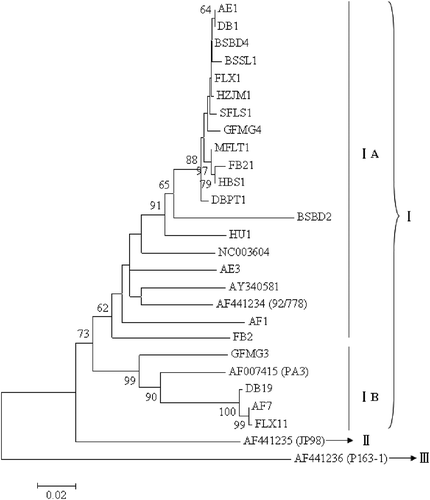
Sequence analysis of CP genes from these isolates revealed that all tested variants clustered into two subgroups IA and IB () represented by isolate 92/778 and PA3 in group I (Galiakparov et al., Citation2003; Goszczynski & Jooste, 2003a). Previous work showed that a GVA isolate LQ58 from Sichuan province in China clustered into group III based on the CP gene sequence (Wang et al., Citation2008). However, in this study, there was no variant clustered into the same branch with representative isolates JP98 and P163-1 in groups II and III, respectively. All 14 isolates contained variants belonging to subgroup IA (). Variants from each isolate clustered into the same subgroup, except for some variants in isolates AF, DB, FLX and GFMG. It was also notable that all the variants from isolates AE, BSBD and FB belonged to GVA-subgroup IA (), although there was a high inter-clone diversity within these isolates (). Variants in subgroup IB were only found in isolate AF, DB, GFMG and FLX with very low frequencies (). We suggest that these variants might have emerged recently, consistent with studies reported by Garcia-Arenal et al. (Citation2001). Viral variants within subgroup IA and subgroup IB had sequence identities ranging from 86.3% to 99.8% and 91.2% to 99.8%, respectively. Sequence identities of variants between two subgroups were 83.8–91.1%.
Fig. 2. Phylogenetic analysis of Grapevine virus A (GVA) coat protein (CP) genes. In total, 24 CP sequences representing different sub-isolates within each of 14 isolates were included in the analysis. Six CP sequences from GenBank (accession no. AF441234, AF441235, AF441236, NC003604, AF007415 and AY340581) were used as references. The tree was constructed with MEGA 4.1 using the neighbour-joining method with 1000 bootstrap replicates. Values below 50% were suppressed. The bar represents 0.02 substitutions per site.
Acknowledgements
This study was financially supported by the Chinese Ministry of Agriculture, Industry Technology Research Project (Grant No. 200903004) and the Ministry of Education of China (Grant No. IRT0548). We are grateful to the scientists C.H. Liu (Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences), L.Z. Gong (Institute of Fruit and Tea, Hubei Academy of Agriculture Sciences) and Y.F. Dong (Xingcheng Fruit Research Institute, Chinese Academy of Agricultural Sciences), who kindly provided grapevine canes.
References
- Angelini , E. , Bertazzon , N. and Borgo , M. 2004 . Diversity among Grapevine leafroll-associated virus 2 isolates detected by heteroduplex mobility assay . J. Phytopathol. , 152 : 416 – 422 .
- Ayllón , M.A. , Lopez , C. , Navas-Castillo , J. , Garnsey , S.M. , Gureei , J. , Flores , R. and Moreno , P. 2001 . Polymorphism of the 5′ terminal region of Citrus tristeza virus (CTV) RNA: incidence of three sequence types in isolates of different origin and pathogenicity . Arch. Virol. , 146 : 27 – 40 .
- Ayllón , M.A. , Rubio , L. , Sentandreu , V. , Moya , A. , Guerri , J. and Moreno , P. 2006 . Variations in two gene sequences of Citrus tristeza virus after host passage . Virus Genes , 32 : 119 – 128 .
- Carstens , E.B. 2010 . Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) . Arch. Virol. , 155 : 133 – 146 .
- Chevalier , S. , Greiff , C. , Clauzel , J.M. , Walter , B. and Fritsch , C. 1995 . Use of immunocapture-polymerase chain reaction procedure for the detection of Grapevine virus A in Kober stem grooving-infected grapevines . J. Phytopathol. , 143 : 369 – 373 .
- Choueiri , E. , Digiaro , M. and Savino , V. 1997 . Further evidence that Grapevine virus A is the agent of Kober stem grooving. Extended Abstracts 12th Meeting ICVG, Lisbon . Portugal , 1997 : 39 – 40 .
- Fattouch , S. , Acheche , H. , M'Hirsi , S. , Mellouli , L. , Bejar , S. , Marrakchi , M. and Marzouki , N. 2005 . RT-PCR–RFLP for genetic diversity analysis of Tunisian Grapevine fanleaf virus isolates in their natural host plants . J. Virol. Methods , 127 : 126 – 132 .
- Fortusini , A. , Scattini , G. , Prati , S. , Cinquanta , S. and Belli , G. 1997 . Transmission of Grapevine leafroll-associated virus 1 (GLRaV-1) and Grapevine virus A (GVA) by scale insects . Proceedings of 12th Meeting of ICVG, Lisbon, Portugal , 1997 : 121 – 122 .
- Galiakparov , N. , Goszczynski , D.E. , Che , X. , Batuman , O. , Bar-Joseph , M. and Mawassi , M. 2003 . Two classes of subgenomic RNA of Grapevine virus A produced by internal controller elements . Virology , 312 : 434 – 448 .
- Garau , R. , Prota , V.A. , Piredda , R. and Prota , U. 1994 . On the possible relationship between Kober stem grooving and Grapevine virus A . Vitis , 33 : 161 – 163 .
- Garau , R. , Prota , V.A. , Boscia , D. , Fiori , M. and Prota , U. 1995 . Pseudococcus affinis Mask., a new vector of Trichovirus A and B . Vitis , 34 : 67 – 68 .
- Garcia-Arenal , F. , Fraile , A. and Malpica , J.M. 2001 . Variability and genetic structure of plant virus populations . Ann. Rev. Phytopathol. , 39 : 157 – 186 .
- Goszczynski , D.E. 2007 . Single-stranded confirmation polymorphism (SSCP), cloning and sequencing reveal a close association between related molecular variants of Grapevine virus A (GVA) and Shiraz disease in South Africa . Plant Pathol. , 56 : 755 – 762 .
- Goszczynski , D.E. and Jooste , A.E.C. 2003a . Identification of divergent variants of Grapevine virus A . Eur. J. Plant Pathol. , 109 : 397 – 403 .
- Goszczynski , D.E. and Jooste , A.E.C. 2003b . Identification of grapevines infected with divergent variants of Grapevine virus A using variant-specific RT-PCR . J. Virol. Methods , 112 : 157 – 164 .
- Goszczynski , D.E. , Preez , J. and Burger , J.T. 2008 . Molecular divergence of Grapevine virus A (GVA) variants associated with Shiraz disease in South Africa . Virus Res. , 138 : 105 – 110 .
- Hommay , G. , Komar , V. , Lemaire , O. and Herrbach , E. 2008 . Grapevine virus A transmission by larvae of Parthenolecanium corni . Eur. J. Plant Pathol. , 121 : 185 – 188 .
- Juke , T.H. and Cantor , C.R. 1969 . “ Evolution of protein molecules ” . In Mammalian protein metabolism , Edited by: Munro , H.N. 21 – 32 . New York : Academic Press .
- Kong , P. , Rubio , L. , Polek , M. and Galk , B.W. 2000 . Population structure and genetic diversity within California Citrus tristeza virus (CTV) isolates . Virus Genes , 21 : 139 – 145 .
- La Notte , P. , Buzkan , N. , Choueiri , E. , Minafra , A. and Martelli , G.P. 1997 . Acquisition and transmission of Grapevine virus A by the mealybug Pseudococcus longispinus . J. Plant Pathol. , 78 : 79 – 85 .
- Mackenzie , D.J. 1997 . “ A standard protocol for the detection of viruses and viroids using a reverse transcription-polymerase chain reaction technique ” . In Document CPHBT-RT-PCR 1.00 , The Canadian Food Inspection Agency .
- Magome , H. , Yoshikawa , N. and Takahashi , T. 1999 . Single–stranded conformation polymorphism analysis of Apple stem grooving capillovirus sequence variants . Phytopathol. , 89 : 136 – 140 .
- Martelli , G.P. 1993 . Graft-transmissible diseases of grapevine. Handbook for Detection and Diagnosis , 263 Rome : FAO Publication Division .
- Martelli , G.P. , Minafra , A. and Sardarelli , P. 1997 . Viti virus, a new genus of plant viruses . Arch. Virol. , 142 : 1929 – 1932 .
- Minafra , A. and Hadidi , A. 1994 . Sensitive detection of Grapevine virus A, B, or leafroll-associated III from viruliferous mealybugs and infected tissue by cDNA amplification . J. Virol. Methods , 47 : 175 – 187 .
- Minafra , A. , Saldarelli , P. and Martelli , G.P. 1997 . Grapevine virus A: nucleotide sequence, genome organization, and relationship in the Trichovirus genus . Arch. Virol. , 142 : 417 – 423 .
- Monette , P.L. and James , D. 1990 . Detection of two strains of Grapevine virus A . Plant Dis. , 74 : 898 – 900 .
- Murolo , S. , Romanazzi , G. , Rowhani , A. , Minafra , A. , La Notte , P. , Branzanti , M.B. and Savino , V. 2008 . Genetic variability and population structure of Grapevine virus A coat protein gene from naturally infected Italian vines . Eur. J. Plant Pathol. , 120 : 137 – 145 .
- Palacio , A. and Duran-Vila , N. 1999 . Single-strand conformation polymorphism (SSCP) analysis as a tool for viroid characterization . J. Virol. Methods , 77 : 27 – 36 .
- Rezaian , M.A. , Krake , L.R. , Cunying , Q. and Hazzalin , C.A. 1991 . Detection of virus-associated dsRNA from leafroll infected grapevines . J. Virol. Methods , 31 : 325 – 333 .
- Ribeiro , G.P. , Saldarelli , P. , Hong , N. , Xiang , B.C. , Zhang , X.L. , Wang , G.P. and Martelli , G.P. 2004 . First record of three grapevine viruses in the Chinese province of Sinkiang . J. Plant Pathol. , 86 : 264
- Rosciglione , B. , Cannizzaro , G. , Castellano , M.A. , Martelli , G.P. and Savino , V. 1983 . Mealybug transmission of Grapevine virus A . Vitis , 22 : 331 – 347 .
- Rosciglione , B. and Castellano , M.A. 1985 . Further evidence that mealybugs can transmit Grapevine virus A (GVA) to herbaceous hosts . Phytopathol. Mediterr. , 24 : 186 – 188 .
- Rubio , L. , Abou-Jawdah , Y. , Lin , H. and Falk , B.W. 2011 . Geographical distant isolates of the crinvirus Cucurbit yellow stunting disorder virus show very low genetic diversity in the coat protein gene . J. Gen. Virol. , 82 : 929 – 933 .
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Mol. Biol. Evol. , 24 : 1596 – 1599 .
- Thompson , J.D. , Gibson , T.J. , Plewniak , F. , Jeanmougin , F. and Higgins , D.G. 1997 . The CLUSTAL_X windows interface:Flexible strategies for multiple sequence alignment aided by quality analysis tools . Nucleic Acids Research , 25 : 4876 – 4882 .
- Turturo , C. , Saldarelli , P. , Dong , Y. , Digiaro , M. , Minafra , A. , Savino , V. and Martelli , G.P. 2005 . Genetic diversity and population structure of Grapevine leafroll-associated virus 3 isolates . J. Gen. Virol. , 86 : 217 – 224 .
- Vigne , E. , Bergdoll , M. , Guyader , S. and Fuchs , M. 2004 . Population structure and genetic variability within isolates of Grapevine fanleaf virus from a naturally infected vineyard in France: evidence for mixed infection and recombination . J. Gen. Virol. , 85 : 2435 – 2445 .
- Wang , G.P. , Hong , N. , Zhang , Z.P. , Zhang , Y. and Jiang , X.F. 1996 . Field survey of viruses on grapevine and stone fruit in Liaoning and Shangdong . China Fruits , 1996 : 39 – 41 .
- Wang , J.H. , Liu , X. , Xi , D.H. , Yuan , S. , Jiang , Y. , Yang , H. , Du , J.B. , Zhang , Z.W. , Chen , K.L. and Lin , H.H. 2008 . Cloning and prokaryotic expression of CP gene of Grapevine virus A Sichuan isolate . Acta Horticulturae Sinica , 35 : 967 – 972 .
- Zorloni , A. , Prati , S. , Bianco , P.A. and Belli , G. 2006 . Transmission of Grapevine virus A and Grapevine leafroll-associated virus 3 by Heliociccus bohemicu . J. Plant Pathol. , 88 : 325 – 328 .