Abstract
Nineteen active bacteriophages against Erwinia amylovora, the causal agent of fire blight, were collected from apple and pear orchards in the Okanagan and Fraser Valleys of British Columbia. Eight survived the isolation, purification and storage processes. Five bacteriophage isolates included in this study lysed more than 50% of the 20 E. amylovora strains tested from BC. Examination by transmission electron microscopy revealed that all eight phages belong to the order Caudovirales, the tailed phages, and included members of the families Myoviridae and Podoviridae. Bacteriophages were characterized by digestion of the phage DNA with four restriction endonucleases and two sets of PCR primers. Two novel groups, RFLP groups 7 and 8, were identified based on differences in restriction fragment patterns. Phages ΦEa1337-26 and ΦEa2345-6 reduced infection by 84% and 96%, respectively, when tested on detached pear blossoms using the epiphyte bacterium Pantoea agglomerans Eh21-5 as a carrier. In addition, bacteriophage ΦEa2345-6, applied in combination with Eh21-5, reduced infection of fire blight on apple flowers of potted apple trees by 56% and compared well with the antibiotic streptomycin.
Résumé
Dix-neuf bacteriophages capables d'infecter Erwinia amylovora, la bactérie causant la brûlure bactérienne, ont été recueillis dans des vergers de pommiers et poiriers dans les vallées de l'Okanagan et Fraser en Colombie-Britannique. Huit ont survécu aux procédés d'isolement, de purification et de conservation. Cinq des phages ont la capacité d'infecter 50% des vingt souches d'E. amylovora de la C.-B. Selon les observations faites par microscopie électronique, les phages appartiennent à l'ordre des Caudovirales, les virus caudés, faisant partie des familles Myoviridae ou Podoviridae. Les phages ont été caractérisés par la digestion avec quatre enzymes de restriction et par amplification par PCR. Deux nouveaux groups ont été identifiés (groupes 7 et 8). Les phages ΦEa1337-26 et ΦEa2345-6 ont réduit le niveau d'infection de 84% et 96% respectivement lors d'essais sur fleurs détachées. De plus, le phage ΦEa2345-6 a réduit le niveau d'infection de la brûlure bactérienne de 56% sur fleurs de pommiers en pot, et a bien rivalisé avec l'antibiotique streptomycine. Pour favoriser la survie des phages in vivo, ils ont été appliqués en association avec la bactérie épiphyte Pantoea agglomerans Eh21-5.
Introduction
Fire blight is an economically important disease of apple, pear and other rosaceous plants caused by Erwinia amylovora (Burrill) Winslow et al. (Jones & Aldwinckle, Citation1990; Bonn & van der Zwet, Citation2000). Most pear and apple cultivars in commercial production in North America are moderately to highly susceptible to fire blight. Control of the disease depends on limiting primary blossom infection in the spring, and rapidly removing infected tissue. Streptomycin is no longer a reliable control material as resistant strains of E. amylovora have been detected in Canada (Sholberg et al., Citation2001) and in several areas of the USA (Jones & Schnabel, Citation2000). Recently, several commercial biopesticides with active ingredients of Pantoea agglomerans strains C9-1 and E325, Pseudomonas fluorescens strain A506, and Bacillus subtilis strain QST 713 have been registered for the control of fire blight in Canada (British Columbia Ministry of Agriculture and Lands, Citation2010). Introduction of these new products has created more options for fruit growers, but variability in efficacy has been reported in British Columbia (BC), Canada (Sholberg & Boulé, Citation2007) and in Michigan, New York and Virginia in the USA (Sundin et al., Citation2009).
The disease control potential of E. amylovora bacteriophages (phages) was first demonstrated in BC by Erskine (Citation1973). In Michigan, Ritchie & Klos (Citation1977) isolated E. amylovora phages from the aerial parts of the apple tree and subsequently described their properties (Ritchie & Klos, Citation1979). Schnabel et al. (Citation1999) reported 26–37% control of fire blight on apple blossoms with a mixture of three phage isolates and demonstrated that high populations of phage were dependent on the presence of E. amylovora. Cost and efficacy considerations make it necessary to apply biopesticides at times when E. amylovora is not yet present on the blossom. To enhance the survival of the phages by supporting replication on blossom surfaces prior to substantial E. amylovora colonization, the epiphytic bacterium Pantoea agglomerans (Ewing & Fife) Gavini et al. Eh21-5, a non-pathogenic relative of E. amylovora (Lehman, Citation2007; Svircev et al., Citation2010) was selected and used in planta as a carrier.
In Michigan, Schnabel & Jones (Citation2001) characterized eight E. amylovora phages by plaque morphology, restriction fragment polymorphisms, PCR analysis, pulsed-field gel electrophoresis and host range studies. Using these techniques along with transmission electron microscopy, Gill et al. (Citation2003) and Lehman (Citation2007) studied 42 and 56 E. amylovora phage isolates, respectively, and assessed their potential as biological control agents (BCAs). Gill et al. (Citation2003) demonstrated that the Ontario phage isolates did not have as complete a host range on the BC isolates of E. amylovora. The BC research team identified local isolates to eventually develop BCAs that could be used effectively by BC fruit growers.
The objective of the present study was to isolate phages of E. amylovora native to BC, characterize them by host range studies, transmission electron microscopy, restriction fragment polymorphisms and PCR analysis, and evaluate their potential as biological control agents against fire blight using P. agglomerans Eh21-5 as a carrier.
Materials and methods
Bacterial strains
Strains of Erwinia amylovora were cultured on 0.8% (w/v) nutrient agar (Difco Laboratories, Detroit, MI) and incubated at 22°C. Isolates were multiplied in nutrient broth (Difco) with constant agitation provided by an orbital shaker. The pathogenicity of E. amylovora strains was established using the green pear test (Billing et al., Citation1960). Strains of bacteria used in this experiment and their origin are listed in . Bacteriophages were plated using the soft agar overlay method described by Adams (Citation1959), in which 100 μL of diluted phage lysate and 100 μL of bacterial host strain (109 CFU mL−1) are added to 3 mL of molten agar (5 g L−1 sucrose, 2.8 g L−1 nutrient broth and 5.2 g L−1 agar), and poured over a bottom layer consisting of 5 g L−1 sucrose, 8 g L−1 nutrient broth and 15 g L−1 agar. Phages were stored in sterile 0.4% (w/v) nutrient broth at 4°C.
Table 1. Origin and sources of Erwinia amylovora (Ea), Pantoea agglomeransa (Eh, Pa) and Pseudomonas fluorescens (Pf) isolates used in this study
Phage isolation and E. amylovora host range analysis
Soil samples for phage isolation were collected twice during the growing seasons of 2004 and 2005 from fruit-growing areas in BC. Composite soil samples (50 g) were gathered at a depth of 10–20 cm within a metre from the base of trees with a history of fire blight (). Soil samples were enriched in liquid cultures with mixtures of E. amylovora strains, as per Gill et al. (Citation2003). The supernatant obtained was plated using the soft agar overlay with each of the six individual E. amylovora hosts used in the enrichment step. Lawns were checked for the formation of plaques after 24 and 48 h. Single plaques were picked from these lawns and placed in 1 mL nutrient broth with 10 μL of chloroform. The tubes were centrifuged at 8000 g for 5 min. The resulting supernatant was transferred to sterile micro-centrifuge tubes and plated using the soft agar overlay method described earlier. Phage isolates were purified by picking single plaques and plating three times for each given plaque morphology. Once picked from a lawn seeded by a given bacterial strain, the phage isolate was propagated and titred on the isolation strain. Phage preparations were diluted to obtain approximately 50 phages per plate.
Table 2. RFLP grouping, origin, morphotype with dimensions, PCR characterization and estimated genome size of the eight BC bacteriophages and phage ΦEa21-4 from Vineland, ON
The soft agar overlay method was used to establish the host range of the eight BC phage isolates. In addition, phage ΦEa21-4 (Lehman et al., Citation2009) was included and grown on BC host Ea2345. An E. amylovora strain was considered ‘sensitive’ to a given phage when the phage infected the host and produced clearly visible plaques within 48 h, ‘not sensitive’ when no plaques were observed and ‘slightly sensitive’ when hazy plaques were produced.
Transmission electron microscopy
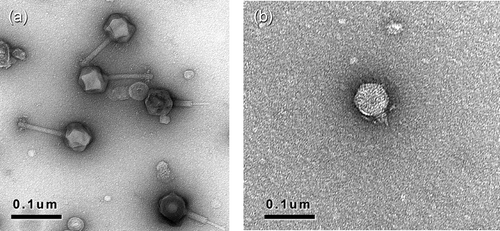
Ten mL of high-titre phage suspensions (109 to 1010 PFU mL−1) were prepared in liquid cultures and centrifuged at 16000 g for 45 min at 4 ° C (Ackermann, Citation2005). The resulting pellet was resuspended in 0.5 mL of Tris–EDTA buffer. One drop of the suspension was placed onto a Formvar film supported by a 300-mesh nickel grid and allowed to sit for 1 min. The excess sample was drawn off by capillary action using a piece of filter paper against the edge of the grid. A drop of 2% uranyl acetate was immediately placed on the grid and the excess stain was drawn off in the same manner. Specimens were viewed by transmission electron microscopy (JEOL 100-CX, Tokyo, Japan) with an accelerating voltage of 80 kV. The electron microscope was calibrated using a carbon grating standard grid. Negatives were taken at a magnification of 20 000×, then digitized at a scale of 1 nm pixel−1. Head and tail (uncontracted) dimensions were measured from enlarged prints on a minimum of 20 phages.
Isolation of phage DNA and molecular analysis
Phage genomic DNA was obtained from high-titre phage isolates grown in liquid culture on their respective host using the cetyltrimethyl ammonium bromide (CTAB) method that was modified from Manfioletti & Schneider (Citation1988). Bacterial nucleic acids were digested by incubating 100 ng of RNase A and 100 U of DNase I with 15 mL of syringe-filtered phage lysate at 22 ° C for 15 min. Nucleases were inactivated and phage particles were lysed by adding 0.8 mL of 0.5 M EDTA (pH 8.0), 0.5 mg of proteinase K (Stratagene, Cedar Creek, TX), and incubation of the mixture at 45 ° C for 15 min. The CTAB: DNA complex was precipitated by adding 440 μL of 5% CTAB in 0.5 M NaCl and cooling the solution on ice for 15 min, followed by centrifugation at 8000 g for 10 min. The resulting pellet was resuspended in 1.2 M NaCl. DNA was precipitated by adding 2 ml of 95% ethanol followed by centrifuging at 8000 g for 10 min. The DNA pellet was washed with 70% ethanol, air-dried and resuspended in 0.5 mL of 10 mM Tris (pH 8.0), and stored at −20°C. DNA concentration was measured with a nanodrop spectrophotometer (ND1000, ThermoFisher Scientific, Nepean, ON).
Phage DNA was digested with EcoRI, BglII, BamHI (Invitrogen, Burlington, ON), or MvnI (Roche Diagnostics, Laval, QC) using 0.2 to 3.2 μg of DNA, 3 units of enzyme, and 0.1 mg mL−1 acetylated bovine serum albumin (New England Biolabs, Pickering, ON) per 50-μL reaction mixture. Samples were digested with either of the enzymes for 3.5 h at 37 ° C. Fragments were separated on a 1% agarose gel in Tris-acetate-EDTA, stained in 1 μg mL−1 of ethidium bromide and visualized using a GelDoc system (Alpha Innotech FC5500, San Leandro, CA). Digestions were repeated twice. Average size of each phage genome was estimated by adding DNA fragments generated by EcoRI restriction enzyme for phages ΦEa1337-26, ΦEa1598-6, ΦEa1598-19, ΦEa2345-6, ΦEa2345-19 and ΦEa21-4.
Phage DNA was amplified with two sets of primers of sequences specific to bacteriophages: primers ‘PEa1A’ and ‘PEa1B’ (Schnabel & Jones, Citation2001), designed to amplify a 304-bp DNA fragment of phage ΦEa1; and ‘Pun45-F’ and ‘Pun45-R’, designed to amplify a 140-bp DNA fragment of phage ΦEa21-4 (D. Roach, personal communication). Primers were synthesized by Integrated DNA Technologies (Coralville, IA). PCR amplification was carried out in 20 μL of reaction mixture containing: 10× buffer (Tris-HCl, 750 mmol L−1; (NH4)2SO4, 200 mmol L−1; and 0.2% v/v Tween® 20; pH 8.8); MgCl2, 2.0 mmol L−1; dNTP 100 μmol L−1; primers, 0.4 μmol L−1; UltraTherm DNA polymerase, 1U (5 U μL−1; BocaScientific, Boca Raton, FL); and DNA sample, 1.5 μL. Reactions were performed in a GeneAmp PCR system 2700 thermocycler (Applied Biosystems, Foster City, CA) with the following amplification conditions: initial denaturation for 3 min at 95°C, followed by 40 cycles (95°C for 20 s, 60°C for 30 s, and 72 for 30 s) and a final extension step of 7 min at 72°C. A PCR reaction mixture lacking DNA was included as a negative control. Reaction products were analyzed on 1.5% (w/v) agarose gels run in 0.5X Tris-borate-EDTA buffer run at 80 V for 55 min, and stained with ethidium bromide. Amplifications were repeated twice.
Assessment of phage control of fire blight in planta
The efficacy of the phages was evaluated on detached pear blossoms (Lehman et al., Citation2008). Newly opened blossoms were harvested from forced pear shoots ‘Bartlett’ and individually placed into scintillation vials such that the peduncle extended through a hole drilled in the lid and into tap water contained in the vial. All treatments contained five blossoms arranged in randomized blocks on racks in closed plastic containers with water at the bottom to keep high humidity levels. In experiment 1, phage ΦEa1337-26 was tested against Erwinia amylovora Ea1337. In experiment 2, ΦEa2345-6, ΦEa2345-19 and ΦEa21-4 were tested against E. amylovora Ea2345. Twenty mL of high-titre phages were prepared in liquid culture as described above and the resulting pellet resuspended in 20 mL of 10 mM sodium phosphate buffer (PB), pH 7.0. Suspensions of Pantoea agglomerans Eh21-5 were prepared from fresh overnight cultures into PB and mixed with phage to obtain a 1:1 ratio of bacteria:phage at 1 × 108 CFU and PFU mL−1. The phages were allowed to multiply on Eh21-5 for 1 h before the suspension was sprayed using a handheld atomizer. Blossoms were allowed to dry for 3 h then were inoculated with a cell suspension of E. amylovora (1 × 106 CFU mL−1) prepared the same way as Eh21-5. Each phage-carrier combination was compared to the standard antibiotic streptomycin at a rate of 0.6 g L−1 (streptomycin sulphate 22.5%, United Agri Products, Dorchester, ON), to the carrier Eh21-5 in PB, and PB alone (negative control). Disease symptoms were scored four to five days after inoculation using the following severity index: 0: no necrosis, blossom healthy; 1: necrosis on the stigma on the hypanthium; 2: necrosis visible on the immediate underside of the blossom; 3: necrosis extends into the ovary, no farther than the widest point; 4: necrosis extends to the base of the ovary; 5: necrosis extends into the peduncle. The severity index of three replications was averaged and the data from three trials (experiment 1) and two trials (experiment 2) were pooled.
In addition, a trial was conducted in a screenhouse on two-year-old ‘Aurora Golden Gala’ apple trees on B9 rootstocks grown in 19-L pots containing soilless media. Each tree was a single replicate and each treatment was replicated five times according to the randomized block design. Phages ΦEa2345-6 and ΦEa2345-19 were grown separately in liquid culture and the lysates were processed by Millipore vacuum filtration (0.2 μm) and concentrated by normal flow diafiltration using an Amicon Model 8400 stirred bell apparatus (Millipore, Oakville, ON) with YM-type regenerated cellulose membrane filters (100 kD) under 3.4 bars of nitrogen pressure until the retentate volume was approximately 150 mL. Dissolved nutrients were partially removed by adding 600 mL of 10 mM PB saline, and again reducing retentate volume to no more than 250 mL. Phage titres in the retentates were assessed by the soft agar overlay method. Pantoea agglomerans Eh21-5 was prepared as described previously and mixed with phages ΦEa2345-6 and ΦEa2345-19 to obtain a 1:1 ratio of bacteria:phage at 1 × 109 CFU and PFU mL−1, respectively.
As in the blossom assay, each phage-carrier mixture was incubated for 1 h and compared to streptomycin, the carrier Eh21-5 in PB, and PB alone (negative control). The suspension was sprayed using a handheld atomizer. Applications took place at 20% bloom and full bloom onto whole trees. Blossoms were inoculated using a backpack sprayer (Solo, Newport News, VA) with a mixture of two isolates of E. amylovora Ea1337 and Ea2345 (6.9 × 108 CFU mL−1 total). These isolates were known to be virulent, streptomycin-sensitive and susceptible to the strains of phages used. Forty-eight hours after the blossoms were inoculated, the trees were wetted for 3 h with overhead sprinklers. Clusters displaying symptoms of fire blight (indicated by blackening of flowers) were recorded eight days following inoculation. Shoots displaying symptoms of fire blight (indicated by blackening and wilting) were recorded 20 days after inoculation. Measurements of the blossom assay and the potted trees trials were subjected to analysis of variance with the General Linear Models Procedure (SAS Institute, Cary, NJ). The data were arcsin transformed and the Waller–Duncan k-ratio t test (k = 100) was used for multiple comparisons of means.
Results
Phage isolation, plaque morphology and host range
Nineteen active phages against E. amylovora were collected from apple and pear orchards in the Okanagan and Fraser Valleys of British Columbia. Eight survived the isolation, purification and storage processes (). Five of the BC phage isolates included in this study lysed more than 50% of the 20 BC E. amylovora strains tested (). E. amylovora strain Ea110, isolated in Michigan, was infected by five of the BC phages. All eight BC phages were able to infect Pantoea agglomerans Eh21-5 but not Pa2092 or Pseudomonas fluorescens Pf1100-6.
Table 3. Host range of eight BC bacteriophages and ΦEa21-4 from Vineland, ON on 20 BC E. amylovora isolates, one E. amylovora from Michigan Ea110, Pantoea agglomerans a Eh21-5 and Pa2092 and Pseudomonas fluorescens Pf1100-6 by using the soft agar overlay method
Transmission electron microscopy
The examination by transmission electron microscopy revealed that the BC phages belong to the order Caudovirales, the tailed phages, and are part of the families Myoviridae or Podoviridae (, ). Particles consist of a head with cubic symmetry and a helical tail The phages of the family Myoviridae have contractile tails consisting of a sheath and a central tube and are identified as morphotype A1 (Ackermann, Citation2001). The Podoviridae attributed to type C1 have short tails.
Molecular analysis
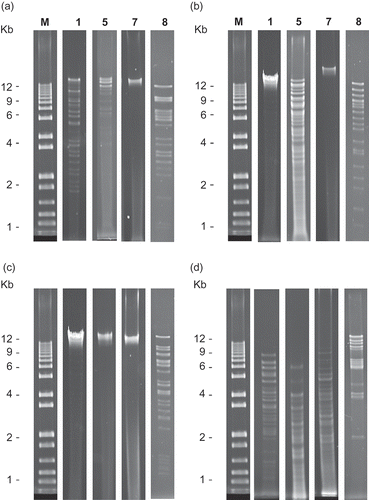
The phages were characterized by digestion of the phage DNA with four restriction endonucleases (). Phage DNA produced unique restriction fragment patterns allowing classification. Three of the BC phages were placed in groups previously described by Gill (Citation2000): RFLP groups 1 and 5, and two novel groups were identified: RFLP groups 7 and 8. ΦEa1615-26 remained ungrouped due to inconsistent digestion or lack of digestion. Three of the BC phages were amplified by the primers specific for phage ΦEa1. The eight BC phages and ΦEa21-4 were amplified by primer Pun45.
Fig. 2. Agarose gel electrophoresis of restriction patterns representative of those exhibited by four phage groups: RFLP groups 1, 5, 7 and 8: a, digestion with EcoRI; b, digestion with BglII; c, digestion with BamHI; and d, digestion with MvnI. Individual lanes are labelled as follows: M: 1 kb Plus DNA ladder (Qiagen), labels 1, 5, 7 and 8 indicating the phage group the pattern represents.
Assessment of phage control of fire blight in planta
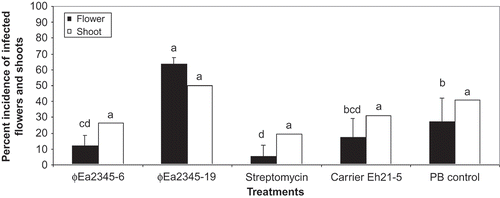
To evaluate their efficacy in planta, the BC phages were tested on detached blossoms (). In experiment 1, phage ΦEa1337-26, applied in combination with carrier Pantoea agglomerans Eh21-5, reduced infection by E. amylovora Ea1337 by 60% compared with the phosphate buffer control. In experiment 2, significantly fewer symptoms of fire blight were observed when inoculated with E. amylovora Ea2345 using phages ΦEa2345-6 and ΦEa2345-19. Phage-carrier combinations ΦEa2345-6-Eh21-5 and ΦEa21-4-Eh21-5 reduced infection by 97% and 88%, respectively. In both experiments, P. agglomerans Eh21-5 alone was also effective. Phage ΦEa2345-6 applied in combination with carrier P. agglomerans Eh21-5 reduced infection of fire blight on apple flowers of potted apple trees by 56% compared with the phosphate buffer control (). Disease development on vegetative shoots followed the same trend as the flower symptoms but no significant difference was observed among treatments.
Fig. 3. Evaluation of phage control of fire blight infection on detached pear blossoms. The rating scale of 0 to 5 was used where 0: no symptoms and 5: infection spread on entire flower surface. In experiment 1, phage ΦEa1337-26 was tested against Erwinia amylovora Ea1337; and in experiment 2, ΦEa2345-6, ΦEa2345-19 and ΦEa21-4 were tested against E. amylovora Ea2345. In both experiments, phages were applied with the carrier Pantoea agglomerans Eh21-5. Each phage carrier combination was compared with streptomycin, Eh21-5 in phosphate buffer (PB), and PB alone (negative control). The severity index of three replications of five detached flowers was averaged and the data from three trials (experiment 1) and two trials (experiment 2) were pooled. Bars of each experiment with the same letters are not significantly different (P ≤ 0.05) according to Waller–Duncan k-ratio (k = 100) t test.
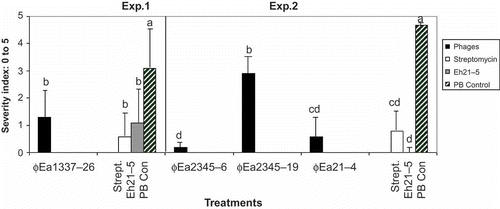
Fig. 4. Per cent incidence of infected flower clusters and vegetative shoots on potted apple trees in a screenhouse treated with ΦEa2345-6 and ΦEa2345-19 and Pantoea agglomerans Eh21-5. The trees were inoculated with a mixture of E. amylovora Ea1337 and Ea2345. Clusters displaying symptoms of fire blight indicated by blackening and wilting were recorded 8 d (flowers) and 20 d (shoots) following inoculation. The data were arcsine transformed for analysis. Bars of the same data set with the same letters are not significantly different (P ≤ 0.05) according to Waller–Duncan k-ratio (k = 100) t test.
Discussion
Five of the eight BC bacteriophages lysed 50% or more of the 20 BC E. amylovora strains tested. This indicates that any of them applied alone or used in combination would likely infect many of the E. amylovora strains present in BC. Gill et al. (Citation2003) reported high lytic activity of phages from RFLP group 1 when tested on five E. amylovora strains from Ontario and four from BC. This is confirmed in the present study since phages ΦEa2345-6 and ΦEa2345-19 from group 1 demonstrated good lytic activity on BC E. amylovora strains as did ΦEa21-4, also belonging to group 1. Gill (Citation2000) found that his group 3 phages, amplified by PEa1-based primers (Schnabel & Jones, Citation2001), showed little or no lytic activity against three of the four E. amylovora strains isolated from BC. The three phages amplified by PEa1 in this study, ΦEa1337-26, ΦEa1594-24 and ΦEa1594-26, showed a weaker lytic activity than other BC phages on BC E. amylovora isolates. The fact that Ea110, isolated in Michigan, was infected by five of the BC phages shows a potential for the BC phages to be effective on E. amylovora strains from other areas. The suitability of Pantoea agglomerans Eh21-5 as a carrier was confirmed with the eight BC bacteriophages able to infect it. Ritchie & Klos (Citation1979) described 11 phages showing a host range limited to E. amylovora and few strains of P. agglomerans (then E. herbicola). Vanneste & Paulin (Citation1990) isolated 10 lytic phages as tools for diagnostic and none were completely specific to E. amylovora. Similarly in this study, of the eight lytic phages isolated in BC, none were completely specific to E. amylovora. However, none of them infected Pseudomonas fluorescens, indicating that they could be used in combination with BlightBan® A506 (Nufarm Americas Inc., Burr Ridge IL), a registered BCA for fire blight that contains P. fluorescens A506 as an active ingredient. Similarities were observed among the phages isolated on the same E. amylovora host but from different locations. Incubation of field samples with a mixture of six E. amylovora host strains was employed specifically to prevent selection of phages based on affinity for any particular bacterial strain and to increase the diversity of the collection. The phages described in this study belong to the virus order Caudovirales as were the E. amylovora phages identified in Ontario (Gill et al., Citation2003; Lehman, Citation2007).
The phages were characterized by digestion of the genomic DNA with four restriction endonucleases. The restriction fragment patterns obtained from phages ΦEa2345-6 and ΦEa2345-19 showed similar patterns to RFLP group 1, and ΦEa1598-6 to group 5 as per Gill (Citation2000). Two novel groups, 7 and 8, were identified based on the production of unique restriction fragment patterns. Group 7 includes ΦEa1594-24 and ΦEa1594-26 producing fragments only when digested with MvnI. ΦEa1337-26 and ΦEa1598-19 produced fragments when digested by the four enzymes, and were placed in group 8. None of the enzymes effectively digested ΦEa1615-26, therefore, it remained ungrouped. Phages with identical restriction patterns can be said to be highly related to each other regardless of site of isolation. Using the primers specific for phage ΦEa1, phages ΦEa1337-26, ΦEa1594-24 and ΦEa1594-26 produced a c. 300 bp PCR product, indicating relatedness to phage ΦEa1 from Michigan. The eight BC phages and ΦEa21-4 were amplified by the Pun45 primers. It would be interesting to see if E. amylovora phages from other areas have this primer sequence in common.
The use of detached blossoms and potted trees instead of assays involving infection of immature pear tissue (Zhao et al., Citation2005) brings efficacy testing closer to field trials, where a candidate BCA is exposed to variable environment conditions, and the existing microbial ecology of the orchard. The results of this study support Lehman's findings (2007) that selected phages, like BC phage ΦEa2345-6, applied in combination with Pantoea agglomerans Eh21-5, can effectively reduce symptom development on blossoms in orchard trees. The efficacy of the carrier alone showed that it contributes directly to the control of E. amylovora populations on the blossom. This epiphyte not only supports the multiplication of the phages but can act as a competitor to E. amylovora (Hattingh et al., Citation1986). A different strain of P. agglomerans is available commercially, as Bloomtime FD (Northwest Agricultural Products, Pasco, WA), and has been shown to inhibit E. amylovora by antibiosis (Pusey et al., Citation2008). The question remains as to why the extra benefit provided by the presence of P. agglomerans does not add to the effectiveness of all the phages, although it may just be an issue of assay sensitivity.
These eight BC phages are of interest since they are the first E. amylovora phages isolated in western Canada. This work contributes to the on-going research in the production of a bacteriophage-based BCA against E. amylovora and provides a basis for the development of local strains of phages to be used by BC fruit growers. The characterization of the eight E. amylovora phages isolated in BC will contribute to a more rapid classification of phages as they are isolated and permits further work on the ecology and distribution of the phages. The challenge of phage-mediated BCAs remains in the transposition of effective control in the laboratory into successful control of fire blight in an orchard setting with variable environment conditions and the existing microbial ecology of the orchard. Phage ecology and the complex bacteriophage–host interactions in the plant environment need to be investigated further.
Acknowledgements
This work was financially supported by Agriculture and Agri-Food Canada. S.M. Lehman was supported by a postgraduate scholarship from the Natural Science and Engineering Research Council of Canada (NSERC). The authors wish to thank Michael Weis for assistance with the transmission electron microscope and the figures, Melanie Walker and Colleen Harlton who provided assistance with molecular techniques and all the summer and co-op students for their assistance.
References
- Ackermann , H.W. 2001 . Frequency of morphological phage descriptions in the year 2000 . Arch. Virol. , 146 : 843 – 857 .
- Ackermann , H.W. 2005 . “ Appendix: Working with bacteriophages: Electron microscopy ” . In Bacteriophages Biology and Applications , Edited by: Kutter , E. and Sulakvelidze , A. 488 – 489 . Boca Raton, FL : CRC Press .
- Adams , M.H. 1959 . Bacteriophages , New York, NY : Interscience Publishers .
- British Columbia Ministry of Agriculture and Lands . 2009 . Integrated Fruit Production Guide 2009 , Victoria, BC : British Columbia Ministry of Agriculture and Lands and British Colombia Fruit Growers’ Association .
- Billing , E. , Crosse , J.E. and Garrett , C.M.E. 1960 . Laboratory diagnosis of fire blight and bacterial blossom blight of pear . Plant Path. , 9 : 19 – 25 .
- Bonn , W.G. and Van Der Zwet , T. 2000 . “ Distribution and economic importance of fire blight ” . In Fire blight: the disease and its causative agent, Erwinia amylovora , Edited by: Vanneste , J.L. 37 – 53 . Wallingford, UK : CAB International .
- Erskine , J.M. 1973 . Characteristics of Erwinia amylovora bacteriophages and its possible role in the epidemiology of fire blight . Can. J. Microbiol. , 19 : 837 – 845 .
- Gill , J.J. 2000 . Bacteriophages of Erwinia amylovora and their potential use in the biological control of fire blight , St. Catharines, ON : Masters thesis, Brock University .
- Gill , J.J. , Svircev , A.M. , Smith , R. and Castle , A.J. 2003 . Bacteriophages of Erwinia amylovora . Appl. Env. Microbiol. , 69 : 2133 – 2138 .
- Hattingh , M.J. , Beer , S.V. and Lawson , E.W. 1986 . Scanning electron microscopy of apple blossoms colonized by Erwinia amylovora and E. herbicola . Phytopathology , 76 : 900 – 904 .
- Jones , A.L. and Aldwinckle , H.S. 1990 . Compendium of apple and pear diseases , 61 – 63 . St. Paul, MN : APS Press .
- Jones , A.L. and Schnabel , E.L. 2000 . “ The development of streptomycin resistant strains of Erwinia amylovora ” . In Fire blight: the disease and its causative agent, Erwinia amylovora , Edited by: Vanneste , J.L. 235 – 251 . Wallingford, UK : CAB International .
- Lehman , S.M. 2007 . Development of a bacteriophage-based biopesticide for fire blight , Catharines, ON : Ph.D. Thesis. Brock University, St .
- Lehman , S.M. , Kim , W.S. , Castle , A.J. and Svircev , A.M. 2008 . Duplex real-time polymerase chain reaction reveals competition between Erwinia amylovora and E. pyrifoliae on pear blossoms . Phytopathology , 98 : 673 – 679 .
- Lehman , S.M. , Kropinski , A.M , Castle , A.J. and Svircev , A.M. 2009 . Complete genome of the broad-host-range Erwinia amylovora phage ΦEa21-4 and its relationship to Salmonella phage Felix O1 . Appl. Environ. Microbiol. , 75 : 2139 – 2147 .
- Manfioletti , G. and Schneider , C. 1988 . A new and fast method for preparing high quality lambda DNA suitable for sequencing . Nucleic Acids Res. , 16 : 2873 – 2884 .
- Pusey , P.L. , Stockwell , V.O. and Rudell , D.R. 2008 . Antibiosis and acidification by Pantoea agglomerans strain E325 may contribute to suppression of Erwinia amylovora . Phytopathology , 98 : 1136 – 1143 .
- Ritchie , D.F. and Klos , E.J. 1977 . Isolation of Erwinia amylovora bacteriophage from aerial parts of apple trees . Phytopathology , 67 : 101 – 104 .
- Ritchie , D.F. and Klos , E.J. 1979 . Some properties of Erwinia amylovora bacteriophages . Phytopathology , 69 : 1078 – 1083 .
- Schnabel , E.L. , Fernando , W.G.D. , Meyer , M.P. , Jones , A.L. and Jackson , L.E. 1999 . “ Bacteriophage of Erwinia amylovora and their potential for biocontrol ” . In VIII International Workshop on Fire Blight. Acta Hortic. Edited by: Momol , M.T. and Saygili , H. Vol. 489 , 649 – 653 .
- Schnabel , E.L. and Jones , A.L. 2001 . Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora . Appl. Env. Microbiol. , 67 : 59 – 64 .
- Sholberg , P.L. , Bedford , K.E. , Haag , P. and Randall , P. 2001 . Survey of Erwinia amylovora isolates from British Columbia for resistance to bactericides and virulence on apple . Can. J. Plant Path. , 23 : 60 – 67 .
- Sholberg , P.L. and Boulé , J. 2007 . “ Efficacy of Apogee, Blightban, Bloomtime, Serenade Max for fire blight control on apple, 2007 ” . In Pest Management Report #54 , Edited by: Labaj , A. 176 – 180 . Guelph, ON : Canadian Phytopathological Society .
- Sundin , G.W. , Werner , N.A. , Yoder , K.S. and Aldwinckle , H.S. 2009 . Field evaluation of biological control of fire blight in the eastern United States . Plant Dis. , 93 : 386 – 394 .
- Svircev , A.M. , Castle , A.J. and Lehman , S.M. 2010 . “ Bacteriophages for control of phytopathogens in food production systems ” . In Bacteriophages in the control of food- and waterborne pathogens , Edited by: Sabour , P.M. and Griffiths , M.W. Washington, DC : ASM Press .
- Vanneste , J.L. and Paulin , J.P. 1990 . Isolation of lytic phages of . Erwinia amylovora. Acta Hortic. , 273 : 95 – 98 .
- Zhao , Y. , Blumer , S.E. and Sundin , G.W. 2005 . Identification of Erwinia amylovora genes induced during infection of immature pear tissue . J. Bacteriol. , 187 : 8088 – 8103 .