Abstract
Prunus and Pyrus species affected with phytoplasma diseases, as well as leafhopper species collected from Prunus and Pyrus fields in Ontario, Canada were tested for presence of phytoplasmas. Preliminary results showed that Graminella nigrifrons is a potential vector for phytoplasma groups 16SrI-W (‘Candidatus Phytoplasma asteris’), and 16SrVII-A (‘Candidatus Phytoplasma fraxini’) to a variety of plant hosts, including peach, apricot, plum and pear. Results showed that G. nigrifrons may be able to transmit both phytoplasma groups simultaneously within the same location and suggest that G. nigrifrons populations appear to have a complex ecology.
Résumé
Des espèces de Prunus et de Pyrus affectées par la phytoplasmose, de même que des espèces de cicadelles collectées dans des champs de Prunus et de Pyrus en Ontario, au Canada, ont été testées pour y déceler des phytoplasmes. Les résultats préliminaires ont montré que Graminella nigrifrons est un vecteur potentiel des groupes de phytoplasmes 16SrI-W (‘Candidatus Phytoplasma asteris’) et 16SrVII-A (‘Candidatus Phytoplasma fraxini’), susceptible de les transmettre à une variété de plantes hôtes, y compris le pêcher, l'abricotier, le prunier et le poirier. Les résultats ont montré que G. nigrifrons peut transmettre simultanément les deux groupes de phytoplasmes en un même endroit, et suggèrent que les populations de G. nigrifrons semblent afficher une écologie complexe.
Introduction
Phytoplasmas are important wall-less non-cultivable prokaryotes in the class Mollicutes. They are obligate symbionts of plants and insects, and in most cases need both hosts for dispersal in nature. In plants, they remain mainly restricted to the phloem tissue and have been recognized by causing diseases in more than 700 plant species (Lee et al., Citation2000; IRPCM, Citation2004), and devastating yield losses in diverse low- and high-value crops worldwide (Bertaccini, Citation2007).
Differentiation and classification of phytoplasmas rely on molecular analyses of conserved genes, in particular the 16S rRNA (Lee et al., Citation1998a; Wei et al., Citation2007, Citation2008). The ribosomal RNA operon became the preferred target for sequencing (Lim & Sears, Citation1989; Lee et al., Citation1998a) and primers have been identified in different positions to amplify phytoplasma-specific fragments from total DNA of infected plants and vectors. Since phytoplasmas occur in low concentrations in the host tissues and their number is subject to seasonal fluctuations, especially in woody hosts, and even the presence of PCR inhibitor compounds in the extracts can vary throughout the year (Marzachi, Citation2006), it is now widely accepted that diagnosis of these pathogens is achieved with a nested PCR approach. A further restriction length polymorphism (RFLP) analysis is often required to achieve the final identification of the pathogen species, even when group-specific primers are used in the nested PCR step (Marzachi, Citation2006). Several hundred phytoplasma strains have been classified on the basis of distinct 16S rRNA gene RFLP patterns resolved on actual and/or virtual electrophoresis gel analysis (Lee et al., Citation2007; Wei et al., Citation2007; Cai et al., Citation2008).
Insect vectors of phytoplasmas are phloem feeders of the Order Hemiptera, mostly leafhoppers (Cicadellidae), planthoppers (Fulgoromorpha) and psyllids (Psyllidae) (Weintraub & Beanland, Citation2006). In insects, phytoplasmas invade the gut and salivary glands and many other tissues, where they can accumulate at great numbers inside and outside cells. Phytoplasmas have to traverse the gut and salivary gland cells in order to reach the saliva for subsequent introduction into the phloem during insect feeding (Hogenhout et al., Citation2008).
Recently, in Canada, eight different phytoplasma groups have been identified associated with diseases in several crop and non-crop species (Olivier et al., Citation2009), including group 16SrI ‘Candidatus Phytoplasma asteris’ (former Aster yellows); 16SrII ‘Candidatus Phytoplasma aurantifolia’ (former Peanut witches' broom); 16SrIII ‘X-disease’; 16SrV ‘Candidatus Phytoplasma ulmi’ (former Elm yellows); 16SrVI ‘Candidatus Phytoplasma trifolii’ (former Clover proliferation); 16SrVII ‘Candidatus Phytoplasma fraxini’ (former Ash yellows); 16SrX-C ‘Candidatus Phytoplasma pyri’ (former Pear decline) and 16SrXII ‘Candidatus Phytoplasma australiense’ (former Stolbur). Macrosteles quadrilineatus (Forbes) is the primary vector of the 16SrI phytoplasma group to numerous plant species in Canada, including cereal, vegetable and fruit crops, ornamentals, herbs and spices (Olivier et al., Citation2009). X-disease is transmitted by at least eight species of leafhoppers, especially Paraphlesius irroratus (Say) and Scaphytopius acutus (Say). The only confirmed vector for the 16SrV group, Scaphoideus luteolus (van Duzee) is not known to occur in Canada. The phytoplasma group 16SrVII is transmitted by the beet leafhoppers, Circulifer tenellus (Baker) and Limottetix Sahlberg sp., and the 16SrX phytoplasma group associated with pear decline is mostly transmitted by Cacopsylla pyricola (Förster) introduced from Europe into the eastern USA in the 1880’s (Olivier et al., Citation2009).
Phytoplasmas of groups 16SrI, 16SrVII (‘Ca. Phytoplasma fraxini’) and 16SrX-C (‘Ca. Phytoplasma pyri’) have been reported to affect Prunus and Pyrus species in Ontario (Wang et al., Citation2008; Hunter et al., Citation2010; Zunnoon-Khan et al., Citation2010a; Zunnoon-Khan et al., Citation2010b). Considering that vector management is the most effective control strategy for phytoplasma diseases (Weintraub & Beanland, Citation2006), surveys to identify potential vector species for Prunus and Pyrus phytoplasma diseases were conducted to assess the importance of insect management in phytoplasma spread.
Materials and methods
Specimen collection
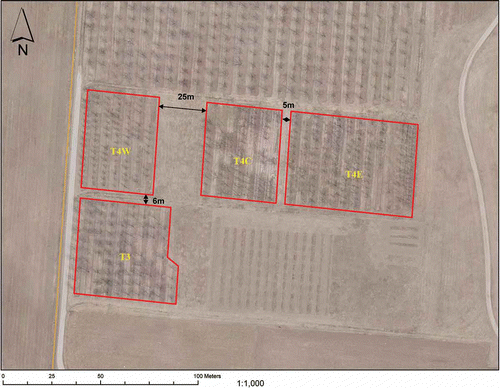
During June-August 2010, over 500 leafhopper specimens were collected using sweep nets in four fields of Prunus and Pyrus at the Canadian Clonal Genebank (CCG): T4-Centre-apricot (T4C), T4-East-peach (T4E), T3-plum and cherry (T3), and T4-West-pear (T4W) (), and subjected to taxonomical identification (Kramer, Citation1967) and PCR testing. Random leaf samples from pear (Pyrus L.), peach (Prunus persica L. Batsch), plum (Prunus domestica L.) and apricot (Prunus armeniaca L.) trees from each field that exhibited symptoms of decline, leaf reddening, witches' broom and rosette-like were also collected and tested ().
Table 1. Phytoplasma groups identified in randomly collected G. nigrifrons specimens and pear, peach, plum and apricot trees from Prunus and Pyrus orchards
DNA extraction and Polymerase Chain Reaction (PCR)
Total DNA was extracted from batches of three leafhopper specimens and 100 mg of leaf midrib tissue (FastDNA Spin kit, MP Biomedicals, USA). Nested PCR was performed using total DNA as template in a PCR reaction with primers specific for the phytoplasma 16S rRNA, R16mF2/R1 (Gundersen & Lee, Citation1996 ) for the first PCR reaction, followed by a nested PCR with either 16S rDNA phytoplasma primers R16F2n/R2 (Gundersen & Lee, Citation1996) or fU5/rU3 (Lorenz et al., Citation1995). For all PCR reactions, 50 ng of the DNA template were added to a 25 μL PCR reaction (Illustra Pure Taq Ready-to-go-PCR-beads, GE Healthcare, UK). For the nested reaction, 1μL of the first round PCR product was used.
Thirty- five cycles were performed for all primer pairs in a DNA Engine Peltier thermal cycler Chromo 4 (Biorad). PCR cycling conditions for both primer pairs R16mF2/R1 and R16F2n/R2 were as follows: 1 min (2 min for the initial denaturation) at 94°C, 2 min at 50°C and 3 min (8 min for the final extension) at 72°C. Five microliters of the PCR products were separated in a 1.5% agarose gel, stained with GelRed Nucleic Acid Stain (Cat 41001, Biotium, Hayward, USA), and visualized with a UV transiluminator in a gel documenter (red, Alpha Innotech, USA).
Cloning, sequencing and sequence analysis
Representative R16F2n/R2 PCR amplicons from both insect and plant samples from each field surveyed were purified (EZNA Cycle Pure kit, Omega Bio-Tek, USA), cloned (pGEM-T Easy Vector, Promega, Madison, USA), and sequenced in both forward and reverse directions (Princess Margaret Hospital, Toronto, Canada). The 16S rDNA sequences were compared with reference sequences in GenBank by BLAST (Altschul et al., Citation1990), including representatives of phytoplasma subgroups 16SrI-W, 16SrI-B, 16SrI-L, 16SrVII-A, 16SrX-A and 16SrX-C, and were compared for sequence similarities (). The 16S rDNA sequences were aligned using Clustal W (Thompson et al., Citation1994) and a phylogenetic tree was constructed using the neighbor-joining method (Saitou & Nei, Citation1987) with the program MEGA version 3.1 (Kumar et al., Citation2004) with default values and 1,000 replicates for bootstrap analysis.
Table 2. Sequence similarities based on the 16S rDNA of phytoplasma isolates identified in Prunus, Pyrus and G. nigrifrons from Ontario compared with those of reference from GenBank
In silico restriction fragment length polymorphism (RFLP)
The trimmed and aligned R16F2n/R2 sequences of the phytoplasmas detected from each field surveyed and those of representatives of phytoplasma subgroups 16SrI-B, 16SrVII-A and 16SrX-C were exported to the in silico restriction analysis and virtual gel plotting program pDRAW32, developed by AcaClone software (http://www.acaclone.com). Each aligned DNA fragment was digested in silico with AluI, BfaI, BstUI, HaeIII, HhaI, HpaII, MseI, RsaI, DdeI, and Tsp509I restriction endonucleases. After in silico restriction digestion, a virtual 3.0% agarose gel electrophoresis image with minimum 50 bp was plotted automatically to the computer screen.
Results
The four fields surveyed yielded specimens of Balclutha impicta (Van Duzee), Delphacodes campestris (renamed Muirodelphax arvensis in 2010), and Graminella nigrifrons (Forbes) (). G. nigrifrons, a member of Membracoidea, Cicadellidae, Deltocephalinae, known as the black-faced leafhopper and widely distributed in North America, including Canada, was the only species that produced PCR amplicons for phytoplasmas, which ranked for 263 out of 322 (81.7%) specimens collected ().
Fig. 2. Graminella nigrifrons, collected in Prunus and Pyrus orchards in Ontario, Canada and identified as the potential vector of phytoplasma groups 16SrI-W and 16SrVII-A.

In silico RFLP and PCR () following sequencing analysis confirmed phytoplasmas were present in plant samples as members of groups 16SrI-W (peach acc. PRU0445, cultivar ‘HW271’; apricot acc. PRU0134, cultivar ‘Harglow’; apricot acc. PRU0142, cultivar ‘Sundrop’; pear acc. PYR0189, cultivar ‘HW614’; pear acc. PYR0190, cultivar ‘HW615’); 16SrVII-A (peach acc. PRU0168, cultivar ‘Harrow Diamond’; peach acc. PRU0176, cultivar ‘Siberian C’; peach acc. PRU0180, cultivar ‘Vanity’; plum acc. PRU0406, cultivar ‘Pembina’); 16SrX-C (peach acc. PRU0336, cultivar ‘Redhaven’; apricot acc. PRU0147, cultivar ‘Wescot’)..
Fig. 3. R16F2n/R2 PCR amplicons representative from phytoplasmas identified in Prunus and Pyrus species and G. nigrifrons in each field. Lane 1: 1 kb molecular marker (MBI Fermentas); Lane 2: PRU0147; Lane 3: G. nigrifrons T4E; Lane 4: PRU0445; Lane 5: G. nigrifrons T4W; Lane 6: PRU0180; Lane 7: Phytoplasma reference control (‘European aster yellows, subgroup 16SrI-B, kindly donated by Prof. A. Bertaccini, University of Bologna); Lane 8: Sterile nuclease-free water (Promega).
BLAST analysis of partial 16S rDNA sequences of representative phytoplasmas detected in Prunus and Pyrus species, and G. nigrifrons samples (GenBank Accession No. JN563606, JN563607, JN563608, JN563609, JN563610) showed over 99% sequence homology with phytoplasma members of groups 16SrI-B, 16SrI-L, 16SrI-W, 16SrX-C, 16SrX-A and 16SrVII-A at NCBI. Sequence homology values were also over 99% when 16S rDNA sequences of phytoplasmas from Prunus, Pyrus and G. nigrifrons were compared with those of phytoplasmas previously reported at the CCG and Ontario province, respectively, in peach (16SrI-W, HQ450211; 16SrVII-A, GU223903) and pear (16SrX-C, GU565959, GU565960). Particularly, for the 16SrX phytoplasma identified in G. nigrifrons, the 16S rDNA sequence shared over 98% sequence homology with that of the phytoplasma reported in pear associated with pear decline in Ontario (16SrX-C, GU565959, GU565960).
Virtual RFLP profiles with all endonucleases corresponding to the 16SrI phytoplasma identified in PRU0445, PRU0134, PYR0190 and the G. nigrifrons in all the fields surveyed were identical to each other. No endonuclease was able to discriminate between the 16SrI phytoplasmas detected in plants and G. nigrifrons and those belonging to subgroups 16SrI-B (AY180943), 16SrI-L (GU223209) or 16SrI-W (HQ450211), except for BstUI that exhibited unique RFLP profiles. This finding supported that 16SrI phytoplasmas detected in PRU0445, PRU0134, PYR0190 and the G. nigrifrons may be members of the 16SrI-W subgroup (). A comparison among the 16S rDNA sequences of these phytoplasmas showed that the 16SrI-W phytoplasmas detected in PRU0445, PRU0134, PYR0190 (JN563606) and the G. nigrifrons (JN563607) shared a common A at the nucleotide position 1096 with the reference 16SrI-W (HQ450211), which differed from 16SrI-L (GU223209) that showed a substitution AxG at the same position ().
Fig. 4a. BstUI (top) and DdeI (bottom) virtual RFLP profiles from 16S rDNA sequences of phytoplasmas identified in Prunus and Pyrus species and G. nigrifrons. MW: MW: jX174DNA-HaeIII molecular marker. Lane 1: PRU0180 (T4E, 16SrVII-A); Lane 2: PRU0445 (T4E, 16SrI-W); Lane 3: PRU0336 (T4E, 16SrX-C); Lane 4: G. nigrifrons (T4E, 16SrX-A); Lane 5: PRU0134 (T4C, 16SrI-W); Lane 6: PRU0147 (T4C, 16SrX-C); Lane 7: G. nigrifrons (T4C, 16SrVII-A); Lane 8: PRU0406 (T3, 16SrVII-A); Lane 9, 10: G. nigrifrons (T3, 16SrI-W, 16SrX-C); Lane 11: PYR0190 (T4W, 16SrI-W); Lane 12: G. nigrifrons (T4W, 16SrI-W); Lane 13: GU223209 (16SrI-L); Lane 14: AF092209 (16SrVII-A); Lane 15: AJ542543 (16SrX-C); Lane 16: AY180943 (16SrI-B); Lane 17: EF392656 (16SrX-A); Lane 18: HQ450211 (16SrI-W).
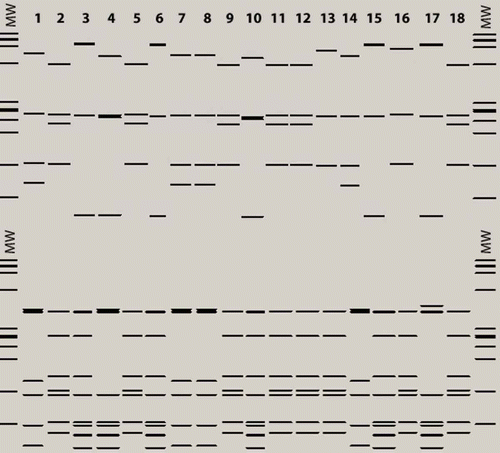
Fig. 4b. AluI (top) and MseI (bottom) virtual RFLP profiles from 16S rDNA sequences of phytoplasmas identified in Prunus and Pyrus species and G. nigrifrons. MW: MW: jX174DNA-HaeIII molecular marker. Lane 1: PRU0180 (T4E, 16SrVII-A); Lane 2: PRU0445 (T4E, 16SrI-W); Lane 3: PRU0336 (T4E, 16SrX-C); Lane 4: G. nigrifrons (T4E, 16SrX-A); Lane 5: PRU0134 (T4C, 16SrI-W); Lane 6: PRU0147 (T4C, 16SrX-C); Lane 7: G. nigrifrons (T4C, 16SrVII-A); Lane 8: PRU0406 (T3, 16SrVII-A); Lane 9, 10: G. nigrifrons (T3, 16SrI-W, 16SrX-C); Lane 11: PYR0190 (T4W, 16SrI-W); Lane 12: G. nigrifrons (T4W, 16SrI-W); Lane 13: GU223209 (16SrI-L); Lane 14: AF092209 (16SrVII-A); Lane 15: AJ542543 (16SrX-C); Lane 16: AY180943 (16SrI-B); Lane 17: EF392656 (16SrX-A); Lane 18: HQ450211 (16SrI-W).
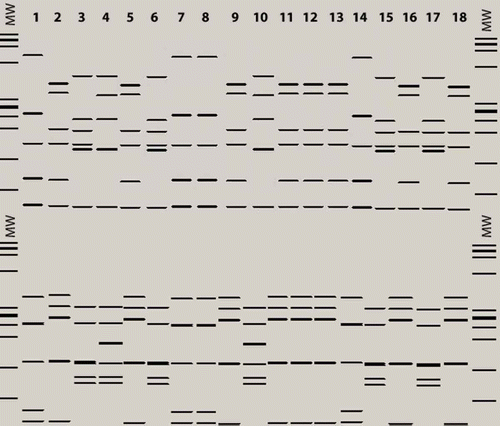
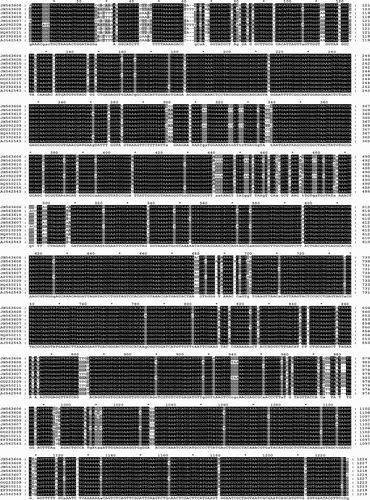
Fig. 5. Sequence alignment based on the 16S rDNA sequences of phytoplasmas identified in Prunus, Pyrus and G. nigrifrons compared with reference phytoplasmas of groups 16SrI, 16SrVII and 16SrX.
The phylogenetic tree () clearly supports RFLP results since the 16S rDNA sequences of the 16SrI phytoplasmas detected in PRU0445, PRU0134, PYR0190 (JN563606) and the G. nigrifrons (JN563607) grouped within the same phylogenetic branch as the phytoplasma reference HQ450211, a member of the new subgroup 16SrI-W recently identified (Arocha-Rosete et al., Citation2011) that affects peach. Sequence similarity analysis confirmed the phylogeny since the PRU0445, PRU0134, PYR0190 phytoplasmas (JN563606) and that detected in G. nigrifrons (JN563607) exhibited the highest (99.8% and 99.4%) of the 16S rDNA sequence identity, respectively, with that of the phytoplasma reference HQ450211 ().
Fig. 6. Phylogenetic tree showing relationships between 16SrI-W, 16SrVII-A and 16SrX-A/16SrX-C phytoplasmas identified in Prunus, Pyrus and G. nigrifrons compared to reference phytoplasmas from Genebank. ‘Ca. P’: ‘Candidatus Phytoplasma’. WB: Witches' Broom. GenBank Accession No.: PRU0134, PYR0190, PRU0445 (JN563606); G. nigrifrons T3 and T4W 16SrI-W (JN563607); PRU0180, PRU0406, G. nigrifrons T4C 16SrVII-A (JN563608); PRU0147, PRU0336 (JN563609); G. nigrifrons T4E and T3 16SrX-A (JN563610).

For the 16SrVII-A phytoplasma identified in PRU0180, PRU0406 and the G. nigrifrons from fields T4E, T4C and T3 (JN563608), the virtual RFLP profiles were all identical to each other and to that of the reference AF092209. The comparison of the 16S rDNA sequences showed that these phytoplasmas share unique features with the reference AF092209 (16SrVII-A), including sequences ‘TTTTT’ and AGGA at positions 60 and 440, respectively; ‘AC’, ‘TAA’ and ACG’ at positions 698, 938 and997, respectively; and ‘T at positions 823, 1058 and 1161 (). Phylogenetic analyses confirmed those RFLP results (). The 16S rDNA sequences of the 16SrVII-A phytoplasma detected in plants and G. nigrifrons group were within the same cluster of the phytoplasma reference AF092209 that belongs to the subgroup 16SrVII-A, and showed a 100% sequence identity with that of the phytoplasma reference AF092209 ().
For the 16SrX phytoplasma identified in PRU0336, PRU0147 and G. nigrifrons from fields T4E, T4C and T3, the virtual RFLP profiles were all identical to those of phytoplasma references AJ542543 (16SrX-C) and EF3922656 (16SrX-A) with all endonucleases, except for MseI and AluI for G. nigrifrons (). The RFLP pattern of the 16SrX phytoplasma detected in G. nigrifrons with DdeI endonuclease showed it to be identical, except for one upper band, to that of the reference EF3922656 (16SrX-A), suggesting that the closest relationship of this phytoplasma is to the subgroup 16SrX-A (). The comparison of the 16S rDNA sequences showed that the phytoplasma identified in G. nigrifrons from fields T4E, T4C and T3 (JN563610) shared unique features (‘A’ at the position 454, and ‘GG’ at position 486) with the reference EF392656 (16SrX-A), which distinguished it from that from plants (JN563609) and the reference AJ542543 (16SrX-C) ().
The phylogenetic tree () showed that the 16S rDNA sequences of the 16SrX phytoplasmas detected in PRU0336, PRU0147 (JN563609) grouped in the same phylogenetic branch of the phytoplasma reference AJ542543 (16SrX-C), supported by 16S rDNA sequence identities of 99.3% and 100%, respectively, with the phytoplasma references EF392656 and AJ542543 (). However, phytoplasmas detected in G. nigrifrons (JN563610) clustered closely with the phytoplasma reference EF392656 (16SrX-A) (). This along with the fact that the 16S rDNA sequence identities of 99.2% and 98.9%, respectively, with those of the phytoplasma references EF392656 and AJ542543 (), suggest that the 16SrX phytoplasma detected in G. nigrifrons may either be a strain more closely related to the subgroup 16SrX-A, or a new 16SrX RFLP variant.
Discussion
RFLP and sequence analyses of the 16S rDNA sequences of phytoplasmas detected in Prunus, Pyrus and G. nigrifrons support that G. nigrifrons may be a potential vector for the 16SrI-W and 16SrVII-A phytoplasmas in all fields surveyed, which are groups previously reported in Ontario (Zunnoon-Khan et al., Citation2010a; Citation2010b; Arocha-Rosete et al., Citation2011). The fact that fields T4E, T4C, T3 and T4W are located close to each other () and that the same 16SrI-W and 16SrVII-A phytoplasmas were found in plants and G. nigrifrons suggests that G. nigrifrons is able to migrate between and around fields efficiently transmitting these two phytoplasmas. However, further transmission trials are required to prove the vector role of G. nigrifrons for both the 16SrI-W and 16SrVII-A phytoplasmas. Since the 16SrX-C and 16SrX-A-related phytoplasmas were individually detected in either the G. nigrifrons or the Prunus species, no conclusions can be established on a potential vector role of G. nigrifrons for the 16SrX-C nor for the 16SrX-A-related phytoplasmas.
It is known that C. pyri and C. pyricola are vectors of high host specificity in transmitting the 16SrX-C group to pear trees (Jensen et al., Citation1964; Carraro et al., Citation1998; Delic et al., Citation2007; Križanac et al., Citation2010), particularly C. pyricola in Canada that transmits pear decline (Olivier et al., Citation2009). However, no Cacopsylla sp. was collected at the time of the surveys at the CCG. The fact that the group 16SrX-C was found in Prunus species suggests that either a non-spotted Cacopsylla-related species could be involved in the transmission of the 16SrX-C phytoplasma to a new Prunus host, or the 16SrX-C phytoplasma may be transmitted by a visitor vector different from G. nigrifrons. Only a 16SrX-A related phytoplasma was found to be carried by G. nigrifrons but not a 16SrX-C phytoplasma; therefore, G. nigrifrons cannot be considered a potential vector for 16SrX-C phytoplasmas. Further investigations on Cacopsylla species will clarify any vector role of these hemipteran, if any, and their possible relationship with either 16SrX-C and/or 16SrX-A-related phytoplasmas in Prunus species.
Multiple phytoplasma infection was found in both plant and insect hosts surveyed in the Prunus, and Pyrus orchards. RFLP and sequencing analyses showed that mixed infections of phytoplasma groups 16SrI-W, 16SrVII-A and 16SrX-C were detected in different peach accessions at the T4E field, while groups 16SrI-W and 16SrX-C were simultaneously detected in apricot at the T4C field. The phytoplasma groups 16SrVII-A and 16SrI-W were found in plum at the T3 field and pear at the T4W field, respectively. G. nigrifrons collected from the field T4E were found to carry a 16SrX-A related phytoplasma, while those collected from fields T4C and T4W were carrying groups 16SrVII-A and 16SrI-W, respectively. G. nigrifrons collected in the field T3 was shown to be able to simultaneously carry two phytoplasma groups, 16SrI-W and 16SrX-A.
Phytoplasmas of groups 16SrI and 16SrVII have been previously reported associated with mixed infections in the field in grapevines in Chile (Fiore et al., Citation2007). In addition, results support previous findings of G. nigrifrons as a vector for group 16SrI since it is the known vector of the maize bushy stunt phytoplasma, ‘Ca. Phytoplasma asteris’ group, subgroup 16SrI-B (Beirne, Citation1956).
It is known that many vectors can transmit more than one type of phytoplasma and that many plants can harbor two or more distinct phytoplasmas, which depends on plant susceptibility to phytoplasma infection, as well as geographic distribution and preferential host(s) of the insect vectors (Lee et al., Citation1998b). G. nigrifrons collected from four adjacent fields on different hosts including peach, apricot, plum and pear have been shown to be able to carry three different phytoplasma groups, 16SrI-W, 16SrVII-A and 16SrX-A-related, and potentially transmit two groups, 16SrI-W and 16SrVII-A.
Plant host range for each phytoplasma in nature is determined largely by the number of natural insect vector species that are capable of transmitting the phytoplasma and by their feeding behaviours (mono-, oligo-, or polyphagus) (Lee et al., Citation1998b; Weintraub & Beanland, Citation2006). So far, more than 75% of all confirmed phytoplasma vector species are monophagous to polyphagous members of Deltocephalinae, and transmit one or more phytoplasma taxa (Weintraub & Beanland, Citation2006). Results suggest that G. nigrifrons is a polyphagous vector with low host specificity and is able to feed on numerous plant species, therefore, contributing to disease spread between nearby fields and showing a very complex ecology.
Results presented show for the first time that apricot, plum and pear are hosts for phytoplasma groups 16SrI-W, 16SrX-C and 16SrVII-A, and peach is a host of phytoplasma group 16SrX-C. Moreover, results present the first record of G. nigrifrons as the potential vector for the 16SrI-W and 16SrVII-A phytoplasma groups on peach, apricot, plum and pear hosts in Ontario. Transmission trials are required to determine G. nigrifrons vector role and ecological factors influencing the transmission of phytoplasma diseases in Prunus and Pyrus. These results have a great impact for the Canadian fruit industry since a new disease management needs to be implemented to halt vector disease spread, and provide more effective control of phytoplasma diseases in Prunus and Pyrus species.
Acknowledgements
We thank Dana Gagnier for assisting with leafhopper collections, Amanda Drouillard (Federal Public Sector Youth Internship Program) for database management, and Dr. Lorna Woodrow for critically reviewing the manuscript; Tom Dufour, Geomatics Technician, Essex region Conservation Authority for the aerial map photo and Tom Murray, Agriculture and Agri-Food Canada, for the photo of G. nigrifrons.
References
- ACACLONE pDRAW32 SOFTWARE http://www.acaclone.com (http://www.acaclone.com)
- Altschul , S. , Gish , W. , Miller , W. , Meyers , E. and Lipman , D. 1990 . Basic local alignment search tool . J. Mol. Biol, , 215 : 403 – 10 .
- Arocha-Rosete , Y. , Zunnoon-Khan , S. , Krukovets , I. , Crosby , W. , Scott , J. , Bertaccini , A. and Michelutti , R. 2011 . Identification and molecular characterization of the phytoplasma associated with peach rosette-like disease at the Canadian Clonal Genebank based on the 16S rRNA gene analysis . Can. J. Plant Pathol , 33 : 127 – 34 .
- Beirne , B.P. 1956 . Leafhoppers (Homoptera: Cicadellidae) of Canada and Alaska . Can. Entomol , LXXXVIII ( Suppl. 2 ) : 1 – 180 .
- Bertaccini , A. 2007 . Phytoplasmas: diversity, taxonomy, and epidemiology . Front. Biosci , 12 : 673 – 689 .
- Cai , H. , Wei , W. , Davis , R.E. , Chen , H. and Zhao , Y. 2008 . Genetic diversity among phytoplasmas infecting Opuntia species: virtual RFLP analysis identifies new subgroups in the peanut witches’ broom phytoplasma group . Int. J. Syst. Evol. Microbiol , 58 : 1448 – 57 .
- Carraro , L. , Loi , N. , Ermacora , P. , Gregoris , A. and Osler , R. 1998 . Transmission of pear decline by using naturally infected . Cacopsylla pyri L. Acta Horticult , 472 : 665 – 668 .
- Delic , D. , Martini , M. , Ermacora , P. , Myrta , A. and Carraro , L. 2007 . Identification of fruit tree phytoplasmas and their vectors in Bosnia and Herzegovina . OEPP/EPPO Bulletin , 37 : 444 – 448 .
- Fiore , N. , Prodan , S. , Paltrinieri , S. , Gajardo , A. , Botti , S. , Pino , A.M. , Montealegre , J. and Bertaccini , A. 2007 . Molecular characterization of phytoplasmas in Chilean grapevines . Bull. Insectol , 60 : 331 – 332 .
- Gundersen , D.E. and Lee , I.M. 1996 . Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs . Phytopathol. Medit , 35 : 144 – 151 .
- Hogenhout , S.A. , Oshima , K. , Ammar , E.D. , Kakizawa , S. , Kingdom , H.N. and Namba , S. 2008 . Phytoplasmas: Bacteria that manipulate plants and insects . Mol. Plant Pathol , 9 : 403 – 423 .
- Hunter , D. , Svircev , M. , Kaviani , M. , Michelutti , R. , Wang , L. and Thompson , D. 2010 . First report of pear decline caused by ‘Candidatus Phytoplasma pyri’ in Ontario, Canada . Plant Dis , 94 : 634
- IRPCM Phytoplasma/Spiroplasma Working Team–Phytoplasma Taxonomy Group . 2004 . ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects . Int. J. Syst. Evol. Microbiol , 54 : 1243 – 1255 .
- Jensen , D.D. , Griggs , W.H. , Gonzales , C.Q. and Schneider , H. 1964 . Pear decline virus transmission by pear psylla . Phytopathology , 54 : 1346 – 1351 .
- Kramer , J.P. 1967 . A taxonomic study of Graminella nigrifrons, a vector of corn stunt disease, and its congeners in the United States (Homoptera: Cicadellidae: Deltocephalinae) . Ann. Entomol. Soc. Amer , 60 : 604 – 616 .
- Križanac , I. , Mikec , Ž. , Budinš , M. , Šeruga , M. and Škori , D. 2010 . Diversity of phytoplasmas infecting fruit trees and their vectors in Croatia . J. Plant Dis. Prot , 117 : 206 – 213 .
- Kumar , S. , Tamura , K. and Nei , M. 2004 . MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment . Brief Bioinform , 5 : 150 – 163 .
- Lee , I-M. , Davis , R.E. and Gundersen-Rindal , D.E. 2000 . Phytoplasma: phytopathogenic mollicutes . Ann. Rev. Microbiol , 54 : 221 – 255 .
- Lee , I-M. , Gundersen-Rindal , D.E. , Davis , R.E. and Bartoszyk , I.M. 1998a . Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences . Int. J. Syst. Evol. Microbiol , 48 : 1153 – 1169 .
- Lee , I-M. , Gundersen-Rindal , D.E. , Davis , R.E. and Bartoszyk , I.M. 1998b . Phytoplasmas: ecology and genomic diversity . Phytopathology , 88 : 1359 – 1366 .
- Lee , I-M. , Zhao , Y. , Davis , R.E. , Wei , W. and Martini , M. 2007 . Prospects of DNA-based systems for differentiation and classification of phytoplasmas . Bull. Insectol , 60 : 239 – 244 .
- Lim , P.O. and Sears , B.B. 1989 . 16S rRNA sequence indicates that plant-pathogenic mycoplasmalike organisms are evolutionarily distinct from animal mycoplasmas . J. Bacteriol , 171 : 5901 – 5906 .
- Lorenz , K.H. , Schneider , B. , Ahrens , U. and Seemüller , E. 1995 . Detection of the apple proliferation and pear decline phytoplasmas by PCR amplification of ribosomal and nonribosomal DNA . Phytopathology , 85 : 771 – 776 .
- Marzachi , C. 2006 . Molecular diagnosis of phytoplasmas . Arab. J. Plant Prot , 24 : 139 – 142 .
- Olivier , C. , Lowery , D.T. and Stobbs , L. 2009 . Phytoplasma diseases and their relationships with insect and plant hosts in Canadian horticultural and field crops . Can. Entomol , 141 : 425 – 462 .
- Saitou , N. and Nei , M. 1987 . The neighbor-joining method: a new method for reconstructing phylogenetic trees . Mol. Biol. Evol , 4 : 406 – 425 .
- Thompson , J.D. , Higgins , D.G. and Gibson , T.J. 1994 . CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice . Nucleic Acids Res , 22 : 4673 – 4680 .
- Wang , L. , Michelutti , R. , Martini , M. , Wang , G. and Hong , N. Survey of ‘Candidatus Phytoplasma’ occurrence in the Canadian Clonal Genebank and commercial plantings . VI International Scientific Seminar on Plant Health and II International Phytoplasma Workshop . Havana , Cuba . 22–26 September .
- Weintraub , P. and Beanland , L. 2006 . Insect vectors of phytoplasmas . Ann. Rev. Entomol , 51 : 91 – 111 .
- Wei , W. , Davis , R.E. , Lee , I-M. and Zhao , Y. 2007 . Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups . Int. J. Syst. Evol. Microbiol , 57 : 1855 – 1867 .
- Wei , W. , Lee , I-M. , Davis , R.E. , Suo , X. and Zhao , Y. 2008 . Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages . Int. J. Syst. Evol. Microbiol , 58 : 2368 – 2377 .
- Zunnoon-Khan , S. , Arocha-Rosete , Y. , Scott , J. , Crosby , W. , Bertaccini , A. and Michelutti , R. 2010a . First report of ‘Candidatus Phytoplasma fraxini’ (group 16SrVII phytoplasma) associated with a peach disease in Canada . New Dis. Reports , 21 : 20
- Zunnoon-Khan , S. , Arocha-Rosete , Y. , Scott , J. , Crosby , W. , Bertaccini , A. and Michelutti , R. 2010b . First report of ‘Candidatus Phytoplasma asteris’-related strain associated with Peach rosette in Canada . Plant Dis , 94 : 916 – 917 .