Abstract
Whole and partial genome sequences are becoming available at an ever-increasing pace. For many plant pathogen systems, we are moving into the era of genome resequencing. The first Phytophthora genomes, P. ramorum and P. sojae, became available in 2004, followed shortly by P. infestans in 2006. Availability of whole genome sequences has provided rapid and immediate advances in several areas also resulting in many practical applications and critical new insights. Availability of comparative genome data facilitated discovery of new classes of effectors, such as the RxLR-dEER and crinkler effector families. Genome data also enabled development of molecular markers for population genomic approaches that provided critical new insights into the evolutionary history of species and clades of Phytophthora. Several select examples of advances resulting from comparative genomic approaches in a concerted effort of the Oomycete research community are reviewed.
Des séquences entières ou partielles de génomes deviennent de plus en plus accessibles. En ce qui concerne plusieurs systèmes plante-agent pathogène, nous abordons l'ère du recéquençage du génome. Les premiers génomes de Phytophthora, P. ramorum et P. sojae, ont été disponibles à partir de 2004, suivis de près par celui de P. infestans en 2006. L'accessibilité à des séquences génomiques entières a favorisé des progrès rapides dans plusieurs domaines, ce qui a engendré des applications pratiques et ouvert de nouvelles perspectives stimulantes. La disponibilité des données comparatives sur le génome a facilité la découverte de nouvelles classes d'effecteurs comme le RxLR-dEER et les familles d'effecteurs crinkler. Les données sur le génome ont aussi permis le développement de marqueurs moléculaires particuliers aux approches en génomique des populations qui fournissent de nouvelles idées séduisantes sur l'histoire évolutive des espèces et des variantes de Phytophthora. Plusieurs exemples typiques de progrès, consécutifs aux approches en génomique comparative et à l'action concertée du milieu de la recherche sur les Oomycètes, sont passés en revue.
Introduction
Plant pathogens in the genus Phytophthora belong to the Oomycota currently classified within the heterokont/chromist clade of the tree of life (Cavalier-Smith & Chao, Citation2006; Riisberg et al., Citation2009). Phytophthora is a genus which is distributed worldwide and affects many if not most commercially grown crops, forest and ornamental species. Phytophthora species such as P. cinnamomi and P. ramorum cause important diseases on hundreds to thousands of forest or landscape plants (Hardham, Citation2005; Grünwald et al., Citation2008). Recently, P. ramorum was found to cause significant disease on a landscape scale on Japanese larch in Europe (Brasier & Webber, Citation2010; Grünwald et al., Citation2012). Other notable Phytophthora species have a small host range, such as P. infestans (Fry, Citation2008) which infects potato and a few other relatives, including tomato and petunia, or P. sojae infecting soybean and some lupines (Tyler, Citation2007). Phytophthora pathogens can be costly to manage and often cause significant crop and postharvest losses on a worldwide scale.
Table 1. Comparison of genome sizes and estimated number of protein-coding genes in Oomycete genomes sequenced to date. The first three Oomycete genomes to be sequenced belonged to the genus Phytophthora
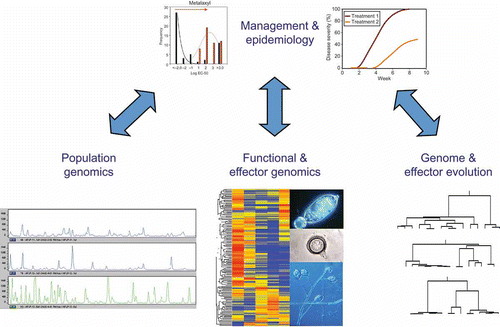
The whole genome sequences of several Phytophthora species have been available for several years (Tyler et al., Citation2006; Haas et al., Citation2009). The availability of these genomes has provided the opportunity to conduct novel research either on a different scale or in novel ways. This review will explore how the availability of Phytophthora whole genome sequences has enabled novel approaches leading to rapid discoveries, new applications and translational research with potential for improving disease management. Availability of whole genome sequences can enable approaches that provide novel tools for population genomic, evolutionary and functional genomic studies that in turn provide insights into a pathogen's plant disease epidemiology and translational tools for better disease management ().
Fig. 1. Illustration of how the availability of whole genome sequences can enable approaches that provide novel tools for population genomic, evolutionary and functional genomic studies that in turn provide insights into a pathogen's plant disease epidemiology and translational tools for better disease management.
Phytophthora genomes
Phytophthora genomes sequenced to date include P. infestans, P. ramorum and P. sojae (Tyler et al., Citation2006; Haas et al., Citation2009). More recently sequenced oomycete genomes include those of Pythium ultimum (Levesque et al., Citation2010) and Hyaloperonospora arabidopsidis (Baxter et al., Citation2010). These oomycete genomes differ drastically in genome size (43–240 Mb), although gene content (15 290–19 027) is roughly equivalent (). Comparative genomic approaches can thus unravel changes in gene arrangement and synteny, changes in gene expression and identify families of genes that underwent gene expansion or contraction. Gene families showing evidence for diversifying selection are likely important for understanding the coevolutionary history of plants and pathogens.
Informative molecular markers and phylogenetic loci
The most immediate applications emerging from the availability of genomes were development of diagnostic, molecular markers particularly for P. ramorum that was previously relatively unknown to science. Martin et al. (Citation2004) developed a PCR protocol based on mitochondrial sequences of the cox I and cox II genes for detection of Phytophthora spp., and discrimination among P. ramorum, P. nemorosa and P. pseudosyringae in particular. Shortly thereafter, the same group developed a real-time PCR assay for detection of P. ramorum (Tooley et al., Citation2006). In parallel, Hayden and colleagues (Citation2004, Citation2006) developed P. ramorum detection methods based on real-time PCR using a nested protocol for the internal transcribed spacer (ITS) region. All these parallel efforts culminated in a large study where 11 reported diagnostic techniques were compared using a standardized DNA library for 315 isolates for over 60 described Phytophthora spp. to develop protocols for routine diagnosis by state and federal agencies in nursery environments (Martin et al., Citation2009).
Another set of diagnostic markers was developed for analysis of populations. Several groups developed simple sequence repeats (SSR) to understand the clonality and structure of populations of P. ramorum. Garnica et al. (Citation2006) mined SSRs from the P. ramorum genome and provided bioinformatically predicted primer sequences. Similarly, Ivors et al. (Citation2006) and Vercauteren et al. (Citation2010, Citation2011) developed and applied SSR markers for P. ramorum while Dorrance & Grünwald (Citation2009) developed SSRs for P. sojae. Finally, Grünwald & Goss (Citation2009) determined frequency and density of SSRs in the genomes of both P. ramorum and P. sojae.
Meanwhile, several groups investigated single nucleotide polymorphisms as diagnostic markers. Bilodeau et al. (Citation2007) discovered SNPs in two genes, β-tubulin and cellulose binding elicitin lectin (CBEL), that can distinguish the European NA1 and the North American EU1 P. ramorum clonal lineages. Analogous work was conducted by Kroon and colleagues (Citation2004) using the cytochrome c oxidase subunit 1 (CoxI) gene to distinguish the same lineages. Similarly, Elliott et al. (Citation2008) used PCR-RFLP to distinguish clonal lineages of P. ramorum.
Another unique effort based on genome availability was provided by Blair and colleagues (2008) who compared the genomes of P. sojae and P. ramorum for sequence loci that are phylogenetically informative at the genus level. They used seven nuclear loci to revise definition of phylogenetic clades in the genus Phytophthora. Availability of genome sequences and the seven loci identified by Blair et al. (Citation2008) also provided opportunity for developing diagnostic websites for the genus Phytophthora: (1) http://www.phytophthoradb.org/ (Park et al., Citation2008), and (2) http://phytophthora-id.org/ (Grünwald et al., Citation2011).
Evolutionary and demographic history
Contemporary analytical tools for the analysis of population genetic variation are powerful resources to infer the evolutionary history of populations of pathogens (Grünwald & Goss, Citation2011). These novel tools allow for the intricate reconstruction of the demographic history of pathogens. Understanding of the evolutionary history of exotic pathogens can, in turn, inform management of emerging epidemics. Plant pathogens in the genus Phytophthora have a long history as exotic and reemerging pathogens that continue to cause significant damage to agriculture and natural ecosystems. Availability of a whole genome sequence for an exotic pathogen such as P. ramorum provided unprecedented opportunities for unravelling the evolutionary history of this pathogen (Grünwald et al., Citation2012).
Table 2. Documented emergence of clonal lineages of the sudden oak death pathogen P. ramorum (Ivors et al., Citation2004, Citation2006; Grünwald et al., Citation2008, Citation2009)
Rapid, genome-enabled discovery of microsatellite markers allowed for extensive demographic analysis of P. ramorum populations. It soon became clear that there exist three distinct clonal lineages named NA1, NA2 and EU1 (Grünwald et al., Citation2009) (). Detailed analysis by several groups has now clearly demonstrated that these three lineages are exotic to both Europe and North America, yet the origin of these lineages remains unknown (Grünwald et al., Citation2008; Goss et al., Citation2009a ; Grünwald & Goss, Citation2009) (). Surprisingly, coalescent analysis and dating of mutations that are lineage-specific versus those that are shared among lineages indicated that the three P. ramorum clonal lineages were anciently diverged, that is they diverged long before the emergence of modern agriculture some 150–400 000 years ago (Goss et al., Citation2009a ). Thus, the three clonal lineages shown in likely were introduced from three separate, reproductively isolated populations.
Fig. 2. Global migration patterns of the sudden oak death pathogen Phytophthora ramorum as supported by research conducted to date. The global population consists of three clonal lineages of unknown origin that were introduced to North America and Europe (Grünwald et al., Citation2008, Citation2009; Goss et al., Citation2009a , Citation2009b , Citation2011; Grünwald & Goss, Citation2009; Vercauteren et al., Citation2010). Clonal lineage EU1 was introduced to Europe and likely moved from Europe to North America (Goss et al., Citation2009a , Citation2011). NA1 was introduced into California (Ivors et al., Citation2006; Mascheretti et al., Citation2008) and NA2 was introduced into either British Columbia, Canada or Washington, US (Goss et al., Citation2011). The source populations for NA1, NA2 and EU1 populations remain unknown and were reproductively isolated long before introduction to Europe or North America (Goss et al., Citation2009a ).
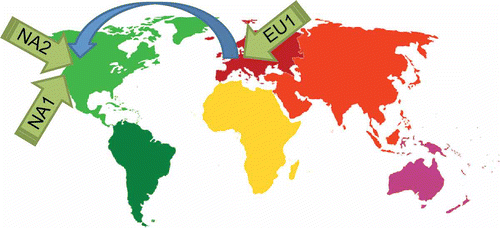
The demographic history of P. ramorum since its emergence is now well documented (Grünwald et al., Citation2012). NA1 established itself in California, USA probably in the early 1990s (Mascheretti et al., Citation2008). Subsequently, the pathogen migrated up (and down) the West coast and West to East via nursery shipments (Prospero et al., Citation2007, Citation2009; Goss et al., Citation2009b ). Recently, the NA1 pathogen migrated among East coast states (California Oak Mortality Taskforce reports).
NA2 in contrast, migrated to the Pacific Northwest and it is currently not clear if it was introduced into British Columbia, Canada or Washington, USA (Ivors et al., Citation2006; Elliott et al., Citation2008; Goss et al., Citation2011).
EU1 was first discovered in Germany (Werres et al., Citation2001) but has since spread throughout Europe (Brasier et al., Citation2004; Grünwald et al., Citation2009; Vercauteren et al., Citation2010). Most recently, this pathogen was shown to cause extensive landscape-level disease on Japanese larch (Brasier & Webber, Citation2010). EU1 was most likely moved from Europe to either British Columbia, Canada, or Washington, USA, by nursery imports from Europe (Goss et al., Citation2011; Grünwald et al., Citation2012).
It has now become clear that P. ramorum has been introduced into North America at least three times, and thus has migrated globally at least four times given the introduction into Europe. All these analyses were critically enabled by availability of whole genome sequences.
Effector discovery
Availability of two simultaneously sequenced Phytophthora genomes was a coup de maître that facilitated comparative genomic approaches and resulted in rapid discovery of new gene families (Tyler et al., Citation2006). The most notable discoveries were made regarding effectors (Kamoun, Citation2006; Tyler, Citation2009). Effectors are small, secreted proteins produced by the pathogen that target host plant molecules and/or alter host plant processes.
One large effector family discovered after availability of whole genome sequences is the RxLR-dEER family. Members of this gene class had been cloned and validated to be avirulence genes corresponding to host R genes, including Avr1b-1 from P. sojae (Shan et al., Citation2004), Avr3a from P. infestans (Armstrong et al., Citation2005) and ATR1 (Rehmany et al., Citation2005) and ATR13 (Allen et al., Citation2004) from H. arabidopsidis. Once genomes were available, it became clear that these four cloned genes all had a conserved gene structure and included the RxLR-dEER domain (Tyler et al., Citation2006). The RxLR-dEER motif is now known to mediate entry of oomycete effectors into host plant cells (Whisson et al., Citation2007; Dou et al., Citation2008b ). The genome sequences of Phytophthora and Hyaloperonospora encode large numbers of genes in several families that have the RxLR domain. For example, P. sojae, P. ramorum and P. infestans have anywhere from 400–600 RxLR effectors depending on the bioinformatic criteria used for mining them out of the available genomes (Tyler et al., Citation2006; Jiang et al., Citation2008). Once the RxLR motif was recognized, rapid progress was made in cloning other RxLR effectors, including identification of Avr4 (van Poppel et al., Citation2008) and AvrBlb1 (Vleeshouwers et al., Citation2008) from P. infestans and Avr4/6 from P. sojae (Dou et al., Citation2008a ). Predicted RxLR effector genes have been used for high throughput screening of germplasm to rapidly identify novel functional host genes (Vleeshouwers et al., Citation2008). This effector-based genomic screening now enables discovery of host genes that promise to accelerate the engineering of Phytophthora-resistant hosts.
Another notable effector group discovered that was genome-enabled is that of the crinklers. Crinkler genes trigger crinkling and necrosis of leaves when overexpressed in Nicotiana benthamiana (Torto et al., Citation2003). Similar to RxLR effectors, crinklers are modular and feature an N-terminal secretory signal and a domain defined by the LxLFLAK amino acid motif (Haas et al., Citation2009).
Conclusions
Availability of whole genome sequences has led to significant, novel discoveries and applications, and thus opened doors for approaches that were previously either inconvenient or cost-prohibitive. Many of the translational results that will emerge out of the genome sequences are still to occur. Immediate applications derived from genome-enabled science occurred mostly in the areas of molecular marker development, population genetics, evolution and effector discovery. The case of P. ramorum exemplifies how this genome provided federal regulatory agencies insights into the migration pathways that led to multiple introductions of this pathogen.
One exciting possibility is that the convergence of decreasing sequencing costs and increased parallel computing resources will provide opportunities for sequencing of all Phytophthora species and resequencing of populations of single species. These approaches might answer long-standing questions about the evolution of sex in the genus, where homothallism and heterothallism are subject to convergent evolution or occasional reversal of traits. Similarly, identification of genes responsible for important traits such as mefenoxam resistance, mating type or host adaptation in Phytophthora will soon be possible, thanks to the novel genomic tools and resources available. Genome-wide association analysis will enable discovery of novel genes responsible for distinct phenotypic traits. Finally, it will further our understanding of what genome features provide narrow host ranges such as those found in P. sojae on Glycine or H. arabidopsidis on Arabidopsis versus broad host ranges such as those found in P. ramorum or Pythium species.
Acknowledgements
I would like to thank all the members of the vibrant Oomycete research community for the wonderful opportunities genomes have provided. Without the collegiality in the Oomycete community, this review would not have been possible. This work was supported in part by funds from USDA-ARS, CRIS Project 5358-22000-034-00D, the USDA-ARS Floriculture Nursery Initiative and the US Forest Service Pacific Southwest Research Station. Mention of trade names or commercial products in this manuscript are solely for the purpose of providing specific information and do not imply recommendation or endorsement by the US Department of Agriculture.
Notes
This paper was a contribution to the symposium entitled ‘Contributions of genomics to plant pathology’ held during the Canadian Phytopathological Society Annual Meeting in Vancouver, British Columbia, June 2010.
References
- Allen , R.L. , Bittner-Eddy , P.D. , Grenville-Briggs , L.J. , Meitz , J.C. , Rehmany , A.P. , Rose , L.E. and Beynon , J.L. 2004 . Host–parasite coevolutionary conflict between Arabidopsis and downy mildew . Science , 306 : 1957 – 1960 .
- Armstrong , M.R. , Whisson , S.C. , Pritchard , L. , Bos , J.I.B. , Venter , E. Avrova , A.O. 2005 . An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm . Proc. Natl. Acad. Sci. USA , 102 : 7766 – 7771 .
- Baxter , L. 2010 . Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome . Science , 330 : 1549 – 1551 .
- Bilodeau , G.J. , Levesque , C.A. , De Cock , A. , Briere , S.C. and Hamelin , R.C. 2007 . Differentiation of European and North American genotypes of Phytophthora ramorum by real-time polymerase chain reaction primer extension . Can. J. Plant Pathol. , 29 : 408 – 420 .
- Blair , J.E. , Coffey , M.D. , Park , S.-Y. , Geiser , D.M. and Kang , S. 2008 . A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences . Fungal Genet. Biol. , 45 : 266 – 277 .
- Brasier , C. and Webber , J. 2010 . Plant pathology: sudden larch death . Nature , 466 : 824 – 825 .
- Brasier , C. , Denman , S. , Brown , A. and Webber , J. 2004 . Sudden oak death (Phytophthora ramorum) discovered on trees in Europe . Mycol. Res. , 108 : 1108 – 1110 .
- Cavalier-Smith , T. and Chao , E.E. 2006 . Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista) . J. Mol. Evol. , 62 : 388 – 420 .
- Dorrance , A. and Grünwald , N.J. 2009 . “ Phytophthora sojae: diversity among and within populations ” . In Oomycete genetics and genomics: biology, interactions, and research tools , Edited by: Lamour , K.H. and Kamoun , S. 197 – 212 . Hoboken , NJ : John Wiley & Sons .
- Dou , D. 2008a . Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b . Plant Cell , 20 : 1118 – 1133 .
- Dou , D. 2008b . RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery . Plant Cell , 20 : 1930 – 1947 .
- Elliott , M. 2008 . PCR-RFLP markers identify three lineages of the North American and European populations of Phytophthora ramorum . For. Pathol. , 39 : 266 – 278 .
- Fry , W.E. 2008 . Phytophthora infestans: the plant (and R gene) destroyer . Mol. Plant Pathol. , 9 : 385 – 402 .
- Garnica , D.P. 2006 . Survey and analysis of microsatellites from transcript sequences in Phytophthora species: frequency, distribution, and potential as markers for the phylum Oomycota . BMC Genomics , 7 : 245
- Goss , E.M. , Carbone , I. and Grünwald , N.J. 2009a . Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum . Mol. Ecol. , 18 : 1161 – 1174 .
- Goss , E.M. , Larsen , M. , Chastagner , G.A. , Givens , D.R. and Grünwald , N.J. 2009b . Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries . PLoS Pathog. , 5 : e1000583
- Goss , E.M. , Larsen , M. , Vercauteren , A. , Werres , S. , Heungens , K. and Grünwald , N.J. 2011 . Phytophthora ramorum in Canada: evidence for migration within North America and from Europe . Phytopathology , 101 : 166 – 171 .
- Grünwald , N. 2011 . Phytophthora-ID.org: a sequence based Phytophthora identification tool . Plant Dis. , 95 : 337 – 342 .
- Grünwald , N.J. and Goss , E.M. 2009 . “ Genetics and evolution of the sudden oak death pathogen Phytophthora ramorum ” . In Oomycete genetics and genomics: biology, interactions, and research tools , Edited by: Lamour , K.H. and Kamoun , S. 179 – 196 . Hoboken , New Jersey : John Wiley & Sons, Inc .
- Grünwald , N.J. and Goss , E.M. 2011 . Evolution and population genetics of exotic and re-emerging pathogens: Novel tools and approaches . Ann. Rev. Phytopathol. , 49 : 249 – 267 .
- Grünwald , N.J. , Garbelotto , M. , Goss , E.M. , Heungens , K. and Prospero , S. 2012 . Emergence of the sudden oak death pathogen Phytophthora ramorum . Trends Microbiol , forthcoming
- Grünwald , N.J. 2009 . Standardizing the nomenclature for clonal lineages of the sudden oak death pathogen, Phytophthora ramorum . Phytopathology , 99 : 792 – 795 .
- Grünwald , N.J. , Goss , E.M. and Press , C.M. 2008 . Phytophthora ramorum: a pathogen with a remarkably wide host-range causing sudden oak death on oaks and ramorum blight on woody ornamentals . Mol. Plant Pathol. , 9 : 729 – 740 .
- Haas , B.J. 2009 . Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature , 461 : 393 – 398 .
- Hardham , A.R. 2005 . Phytophthora cinnamomi . Mol. Plant Pathol. , 6 : 589 – 604 .
- Hayden , K. , Ivors , K. , Wilkinson , C. and Garbelotto , M. 2006 . TaqMan chemistry for Phytophthora ramorum detection and quantification, with a comparison of diagnostic methods . Phytopathology , 96 : 846 – 854 .
- Hayden , K.J. , Rizzo , D. , Tse , J. and Garbelotto , M. 2004 . Detection and quantification of Phytophthora ramorum from California forests using a real-time polymerase chain reaction assay . Phytopathology , 94 : 1075 – 1083 .
- Ivors , K. 2006 . Microsatellite markers identify three lineages of Phytophthora ramorum in US nurseries, yet single lineages in US forest and European nursery populations . Mol. Ecol. , 15 : 1493 – 1505 .
- Ivors , K.L. , Hayden , K.J. , Bonants , P.J.M. , Rizzo , D.M. and Garbelotto , M. 2004 . AFLP and phylogenetic analyses of North American and European populations of Phytophthora ramorum . Mycol. Res. , 108 : 378 – 392 .
- Jiang , R.H.Y. , Tripathy , S. , Govers , F. and Tyler , B.M. 2008 . RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members . Proc. Natl. Acad. Sci. USA , 105 : 4874 – 4879 .
- Kamoun , S. 2006 . A catalogue of the effector secretome of plant pathogenic oomycetes . Ann. Rev. Phytopathol. , 44 : 41 – 60 .
- Kroon , L.P.N.M. , Verstappen , E.C.P. , Kox , L.F.F. , Flier , W.G. and Bonants , P.J.M. 2004 . A rapid diagnostic test to distinguish between American and European populations of Phytophthora ramorum . Phytopathology , 94 : 613 – 620 .
- Levesque , C.A. 2010 . Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire . Genome Biol. , 11 : R73
- Martin , F.N. 2009 . Evaluation of molecular markers for Phytophthora ramorum detection and identification: testing for specificity using a standardized library of isolates . Phytopathology , 99 : 390 – 403 .
- Martin , F.N. , Tooley , P.W. and Blomquist , C. 2004 . Molecular detection of Phytophthora ramorum, the causal agent of sudden oak death in California, and two additional species commonly recovered from diseased plant material . Phytopathology , 94 : 621 – 631 .
- Mascheretti , S. , Croucher , P.J.P. , Vettraino , A. , Prospero , S. and Garbelotto , M. 2008 . Reconstruction of the sudden oak death epidemic in California through microsatellite analysis of the pathogen Phytophthora ramorum . Mol. Ecol. , 17 : 2755 – 2768 .
- Park , J. 2008 . Phytophthora database: a forensic database supporting the identification and monitoring of Phytophthora . Plant Dis. , 92 : 966 – 972 .
- Prospero , S. , Grünwald , N.J. , Winton , L.M. and Hansen , E.M. 2009 . Migration patterns of the emerging plant pathogen Phytophthora ramorum on the west coast of the United States of America . Phytopathology , 99 : 739 – 749 .
- Prospero , S. , Hansen , E.M. , Grünwald , N.J. and Winton , L.M. 2007 . Population dynamics of the sudden oak death pathogen Phytophthora ramorum in Oregon from 2001 to 2004 . Mol. Ecol. , 16 : 2958 – 2973 .
- Rehmany , A.P. 2005 . Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines . Plant Cell , 17 : 1839 – 1850 .
- Riisberg , I. 2009 . Seven gene phylogeny of heterokonts . Protist , 160 : 191 – 204 .
- Rizzo , D.M. , Garbelotto , M. , Davidson , J.M. , Slaughter , G.W. and Koike , S.T. 2002 . Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California . Plant Dis. , 86 : 205 – 214 .
- Shan , W. , Cao , M. , Leung , D. and Tyler , B.M. 2004 . The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b . Mol. Plant– Microbe Interact. , 17 : 394 – 403 .
- Tooley , P.W. , Martin , F.N. , Carras , M.M. and Frederick , R.D. 2006 . Real-time fluorescent polymerase chain reaction detection of Phytophthora ramorum and Phytophthora pseudosyringae using mitochondrial gene regions . Phytopathology , 96 : 336 – 345 .
- Torto , T.A. 2003 . EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. , 13 : 1675 – 1685 .
- Tyler , B.M. 2007 . Phytophthora sojae: root rot pathogen of soybean and model oomycete . Mol. Plant Pathol. , 8 : 1 – 8 .
- Tyler , B.M. 2009 . Entering and breaking: virulence effector proteins of oomycete plant pathogens . Cell. Microbiol. , 11 : 13 – 20 .
- Tyler , B.M. 2006 . Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis . Science , 313 : 1261 – 1266 .
- Van Poppel , P.M. 2008 . The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector . Mol. Plant– Microbe Interact. , 21 : 1460 – 1470 .
- Vercauteren , A. 2010 . Clonal expansion of the Belgian Phytophthora ramorum populations based on new microsatellite markers . Mol. Ecol. , 19 : 92 – 107 .
- Vercauteren , A. , Larsen , M. , Goss , E.M. , Grünwald , N.J. , Maes , M. and Heungens , K. 2011 . Identification of new polymorphic microsatellite markers in the NA1 and NA2 lineages of Phytophthora ramorum . Mycologia , 103 : 1245 – 1249 .
- Vleeshouwers , V.G. 2008 . Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes . PLoS One , 3 : e2875
- Werres , S. and De Merlier , D. 2003 . First detection of Phytophthora ramorum mating type A2 in Europe . Plant Dis. , 87 : 1266
- Werres , S. 2001 . Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum . Mycol. Res. , 105 : 1155 – 1165 .
- Whisson , S.C. 2007 . A translocation signal for delivery of oomycete effector proteins into host plant cells . Nature , 450 : 115 – 118 .