Abstract
Organic amendments influence chemical and microbial compositions in soils and also susceptibility to plant diseases. The purpose of this study was to establish different parameters that interfere with pathogen growth in soil. Four different organic-amendment regimes, i.e. slurry, compost, slurry-dung and compost-slurry-dung, were applied to two fields proximate to each other on the same farm and covered with the same grass-clover ley in the last two years preceding sampling. Before that period, there were differences in cropping and management practices between both fields. Chemical analyses of the soils revealed no differences between organic amendments, whereas significant differences were present between fields in all C, N and pH values. Growth of R. solani AG2-2IIIB in soil, measured by damping-off in sugar beet plants, was influenced by the interaction of organic amendment with field type. Diversity and evenness values of the microbial communities, studied by PCR-DGGEs specific for bacteria, fungi, Pseudomonas and ammonia-oxidizing β-proteobacteria, revealed different patterns, i.e. no differences between organic amendments, but clear differences between fields. Multivariate analyses done on individual species of the four groups, as represented by band location and size in PCR-DGGE fingerprints, and by inclusion of chemical and R. solani AG2-2IIIB growth parameters as ‘environmental’ variables, revealed strong and occasionally significant effects of organic matter content, water-dissolvable organic carbon and pH on microbial communities. It was therefore concluded that different organic amendments had different effects on pathogen growth in soils of both fields and that organic matter content and pH influenced soil microbial compositions most.
Résumé
Les amendements organiques influencent la composition chimique et microbienne des sols ainsi que la sensibilité des plantes aux maladies. Le but de cette étude était de définir divers paramètres qui entravent la croissance des agents pathogènes dans le sol. Quatre différentes formules d'amendements organiques, lisier, compost, lisier et fumier, compost, lisier et fumier, ont été appliquées dans deux champs proches l'un de l'autre, sur la même ferme, champs qui ont été ensemencés pendant deux ans avant l'échantillonnage avec le même mélange graminées et trèfle pour prairie artificielle. Auparavant, les deux champs étaient cultivés et gérés différemment. Les analyses chimiques des sols n'ont indiqué aucune différence entre les amendements organiques, tandis qu'il y avait des différences marquées entre les champs quant aux teneurs en carbone (C) et en azote (N2) ainsi qu'aux valeurs de pH. La croissance de Rhizoctonia solani AG2-2IIIB dans le sol, évaluée en fonction de la fonte des semis chez la betterave à sucre, a été influencée par les interactions des amendements organiques avec le type de champ. La variété et l'uniformité de répartition des communautés microbiennes, analysées par PCR-DGGE spécifique des bactéries, des champignons, des Pseudomonas et des β-protéobactéries oxydatrices d'ammoniac, ont révélé des modes distincts, c'est-à-dire qu'il n'y avait aucune différence entre les amendements, mais des différences notables entre les champs. Les analyses multivariables effectuées sur des espèces particulières des quatre groupes, telles qu'elles étaient représentées par la localisation et les dimensions des bandes des empreintes génétiques obtenues par PCR-DGGE ainsi que par l'inclusion des paramètres chimiques et de croissance de R. solani AG2-2IIIB en tant que variables « environnementales », ont révélé des effets marqués et, à l'occasion, significatifs du contenu de la matière organique, du carbone organique hydrosoluble et du pH sur les communautés microbiennes. On en a par conséquent conclu que différents amendements organiques avaient des effets différents sur la croissance des agents pathogènes dans le sol des deux champs et que le contenu de la matière organique ainsi que le pH influençaient le plus les compositions microbiennes du sol.
Introduction
Microbial community composition of soils is highly complex, and locally differs depending on soil type, land use and plant growth. The structure of microbial communities in agricultural soils is generally governed by soil treatments like tillage (Doran, Citation2011), amendments with different kinds of organic materials (Hoitink & Boehm, Citation1999; Pascual et al., Citation2002; Gorissen et al., Citation2004; Pérez-Piqueres et al., Citation2006), chemical fertilizers (Sarniguet et al., Citation1992a, Citation1992b) and crop growth (Kennedy & Smith, Citation1995; Mazzola, Citation2002; Garbeva et al., Citation2006). Agricultural management practices can exert different effects on microbial activities and plant disease control in soils (Larkin et al., Citation2011). Suppression of plant diseases is an important function of arable soils, but it is difficult to find microbiological parameters that report on antagonism towards phytopathogens in soil (Janvier et al., Citation2007). Because phytopathogens are integral components of natural soil communities, severe disease outbreaks in agricultural fields can be regarded as indicators for ecosystem distress (Van Bruggen & Semenov, Citation2000). Nutrient availability is an important criterion for soil-borne facultative saprotrophic phytopathogens to become prevalent in soils and their possible outgrowth can be constrained by soil-indigenous micro-organisms competing for the same nutrients (Hoitink & Boehm, Citation1999; Grünwald et al., Citation2000; Termorshuizen et al., Citation2006).
Most important microbial groups found to be responsive to various soil treatments and crop rotations were: Pseudomonads (Sarniguet et al., Citation1992a, 1992b; Hoitink & Boehm, Citation1999; Mazzola, Citation2002; Garbeva et al., Citation2006), fungi (Smit et al., Citation1999; Mazzola, Citation2002; Pascual et al., Citation2002; Anderson & Cairney, Citation2004), Lysobacter (van Overbeek & van Elsas, Citation2008, Postma et al., Citation2010) and Burkholderia species (Salles et al., Citation2004), and α- and β-proteobacteria (Schönfeld et al, Citation2003; Senechkin et al., Citation2010). Members of these groups are often associated with the suppression of phytopathogens in soils and it is generally assumed that these groups play important roles in disease suppression.
The purpose of this study was to find correlations between chemical and microbiological parameters in soil and susceptibility to a typical soilborne phytopathogen like Rhizoctonia solani Kuhn. Therefore, we applied four different organic amendments to the same soil in two adjacent fields with identical grass-clover leys in the 2 years preceding soil sampling. The organic amendments are realistic for practice and differed from each other in quality and quantity of total organic carbon and nitrogen input. We determined chemical and microbiological parameters in the different plots, the last by virtue of group-specific molecular fingerprinting on soil DNA extracts. Fungi and Pseudomonas were considered to be microbial groups involved in antagonism towards R. solani AG3 (Garbeva et al., Citation2006), whereas chemolitho-autotrophic ammonia oxidizing β-proteobacteria (Kowalchuk et al., Citation2000; Kowalchuk & Stephen, Citation2001) were considered to be bacterial groups responsive to differences in available nitrogen. Acquired information will be relevant for the control of plant diseases caused by soilborne pathogens in sustainable agriculture.
Materials and methods
Origin of the soils
Fields, denoted as 1 and 6, were located at the Droevendaal experimental farm, Wageningen, the Netherlands (coordinates, 5° 39′ 36.69′′, 51° 59′30.30′′). The farm was converted to organic farming in 2002 and accredited as an ‘organic farm’ in 2004. Before 2002, field 1 was cultivated by crop rotation (rye, potato, grass-clover) and managed under conventional farming practices, whereas field 6 was covered with grass and managed under organic agricultural practices. From 2002 onwards, four replicate plots (8 × 9 m in area) in randomized order, were established over both fields (each with a total area of 36 × 28 m) with the following treatments: liquid cattle manure, slurry (S), green waste compost (C), combination of slurry and solid cattle manure, dung (SD), and combination of compost, slurry and dung (CSD) (for details see ). During this period, plots in both fields were either covered with a grass-clover ley for 4 years (field 1) or cropped with rye (2002), potato (2003) and then covered with a grass-clover ley for 2 years (field 6) (). In September 2005, mixed samples were taken at two locations per plot. Each sample consisted of soil from four bore cores (10 cm in diameter), taken within an area of 40 × 40 cm to a depth of 20 cm each. Samples within the same plot were taken at a distance of 6 m from each other along the diagonal transect of the plot, keeping a minimal distance of 2 m from the borders of the plot. A total of 32 samples were thus taken and soils were stored for not longer than 1 week at 15 °C. Chemical analyses were applied on all soil samples as described in Senechkin et al. (Citation2010), with the exception of the organic matter (OM) content which was performed as described in Ball (Citation1964).
Table 1. History and management of the field soils from experimental farm ‘De Droevendaal’
Soil DNA extraction, PCR amplification and denaturing gradient gel electrophoresis
Total DNA was extracted from all soil samples using the MoBio Ultraclean soil DNA extraction kit (MO Bio Laboratories, Biozym TC, Landgraaf, the Netherlands). Therefore, 0.5 mL of sodium pyrophosphate solution containing 0.5 g soil and 50 mg of glass beads were added to microtubes and cells were dislodged from soil particles by bead beating (Hybaid Ribolyser, Hybaid, Middlesex, UK) for 60 s. Cell lysis and DNA purification was performed according to the protocol provided by the manufacturer. An additional DNA purification step was performed using the Wizard DNA clean-up kit (Promega, Leiden, the Netherlands). DNA quality was checked upon electrophoresis in 0.8% agarose gels stained with ethidium bromide.
Bacterial PCR amplification with primers F968 with GC clamp (Muyzer et al., Citation1993) and R1378 (Heuer et al., Citation1997) was performed according to Van Elsas & Wolters (Citation1995). Pseudomonas-specific PCR amplification was performed with the primers PsF and PsR (Widmer et al., Citation1998) in the first, and with PsR and F968GC in the second step, according to the procedure described in Garbeva et al. (Citation2004). Ammonia-oxidizing β-proteobacteria (AOB) PCR amplification with CTO189f-GC and CTO654r primers was performed according to Kowalchuk et al. (Citation1997). Fungal PCR amplification was performed using primers ITS4 (White et al., Citation1990) and EF4 (Smit et al., Citation1999) in a first PCR, and primers ITS1-F (Gardes & Bruns, Citation1993) with GC clamp and ITS2 (White et al., Citation1990) in a second, nested, PCR according to the procedure described in Anderson & Cairney (Citation2004).
For DGGE, polyacrylamide gels (6%) were prepared with denaturing gradients of 45–65% (100% denaturant consists of 7M urea and 40% formamide) for total bacterial-, Pseudomonas- and AOB-DGGEs and of 30–80% for total fungal DGGE. Gels were loaded with 15-μL volumes prepared by mixing 10 μL PCR product (c. 200 ng) with 5 μL loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF and 30% glycerol). All gels were placed in a PhorU2 apparatus (Ingeny, Goes, NL) set at 60 °C and run at 100 V for 16 h. Then, gels were stained with SYBR Gold (Molecular Probes, Leiden, NL) and digitized fingerprints from individual lanes were normalized using a marker loaded at three different positions in the gel with Molecular Analyst software (version 1.61, BioRad, Veenendaal, NL).
Plant disease development tests
Spread of Rhizoctonia solani AG2-2IIIB in soil was recorded as damping-off in sugar beet (Beta vulgaris ‘Aligator’) seedlings growing at different positions from the R. solani AG2-2IIIB-point-of-inoculation in soils of all 32 samples, according to the experimental design described in Postma et al. (Citation2008). In short, 64 trays of 4 × 25 × 30 cm in size were filled with each of the 32 soils in duplicate, one for treatment with R. solani AG2-2IIIB and the other served as untreated control. Sugar beet seeds were planted in two rows of 11 seeds, keeping a distance of 20 mm between each seed in a row. After seedling emergence (after approximately 7 d), soils were treated by placing oat kernels with R. solani AG2-2IIIB 20 mm in front of the first seedling in each row (treatment with R. solani AG2-2IIIB), or soil in trays were kept untreated (non-inoculated control). Then, treated and control trays were checked for damping-off in sugar beet seedlings at days 7, 14, 21 and 25. Numbers of diseased plants from the two rows per tray were added and expressed as percentage of total number of plants per tray (22). From these data, the ‘area under the disease progress curve’ (AUDPC) and the total percentage of diseased plants after 25 d (cumulative disease incidence, DI) were calculated.
Experimental design and statistical analysis
The experimental fields were designed according to a split-plot scheme, consisting of two fields, separated into two blocks, each consisting of four plots (Senechkin et al., Citation2010). One treatment was applied for each plot and two samples per plot were independently analyzed. Values averaged from duplicate samples of each plot of chemical, microbiological evenness and diversity, and R. solani AG2-2IIIB growth parameters were used for making comparisons between organic amendment and field types. For multivariate analysis, values of all 32 samples were separately taken into consideration.
Distance matrices of PCR-DGGE banding patterns were calculated with GelcomparII software (version 4.5; Applied Maths, Woluwe, Belgium) using Pearson correlation, and clustering was performed using the unweighted-paired group mathematical averages (UPGMA) algorithm. Location and relative intensity of individual bands in fingerprints were taken into account, assuming that these represent single populations within each community. The correlation was calculated using Dice coefficient of similarity and a relatedness tree was produced with the algorithm of the Molecular Analyst software.
Multivariate analysis on PCR-DGGE fingerprints was performed using CANOCO 4.53 (Biometris, Wageningen University and Research Centre, NL). Hill Evenness (E) and Shannon diversity (H′) values were calculated from the fingerprints made with the four different microbial-group-specific primer systems. Bands in PCR-DGGE fingerprints were used as ‘species’ variables, whereas nominal values for organic amendment and field, and quantitative values for all chemical and R. solani AG2-2IIIB growth parameters (AUDPC and DI) were used as ‘environmental’ variables. Indirect gradient analysis between variables was performed by correspondence analysis (CA) and direct gradient analysis by redundancy analysis (RDA). In order to judge for significance of effects of environmental parameters on species compositions, a Monte Carlo permutation test based on 499 permutations, was included. Effects were considered to be significant at levels of P ≤ 0.05, whereas trends in effects were considered to be present at P levels of between 0.05 and 0.1.
Significance of differences between average E and H′ values of total bacteria, fungi, Pseudomonas and AOB, and of AUDPC and non-transformed and arcsine-transformed DI values for R. solani AG2-2IIIB growth, were calculated by ANOVA (Genstat 10th edition, Rothamsted Experimental Station, Harpenden, UK). Differences were considered to be significant at levels of P ≤ 0.05.
Results
Effect of organic amendment and field types on the chemical composition of soil
The average values of all chemical parameters calculated per organic amendment for each of the two fields were published before (Senechkin et al., Citation2010), except for the organic matter fraction in the different soil samples. No significant differences between the four organic amendments over the two fields were found for all six chemical parameters. When the data of the two fields were combined, significant differences between organic amendments were found only for OM, i.e. the percentage OM was lowest in CSD (2.90) and similar for S (3.12), C (3.12) and SD (3.29). When the data of all organic amendment treatments were combined per field, there were significant differences between both fields for all six parameters. The highest values in field 1 were found for pH (4.86 in field 1 and 4.53 in field 6) and NO3-N (0.23 g kg−1 in field 1 and 0.184 g kg−1 in field 6), whereas the highest values in field 6 were found for OM (2.49% in field 1 and 3.71% in field 6), water-dissolvable organic carbon (DOC) (71 mg kg−1 in field 1 and 95 mg kg−1 in field 6), total nitrogen (Ntot) (1.28 g kg−1 in field 1 and 1.57 g kg−1 in field 6) and total carbon (Ctot) (10.6 g kg−1 in field 1 and 17.0 g kg−1 in field 6).
Effect of organic amendment and field types on the microbiological composition of soil
Bacterial, fungal, Pseudomonas and AOB PCR-DGGEs, performed on soils of all 32 soil samples revealed the presence of between 34 and 51 bands in bacterial, 34 and 48 bands in fungal, 10 and 33 bands in Pseudomonas, and 34 and 51 bands in AOB fingerprints. Dendrograms constructed on the basis of intensity and position of these bands in the gels revealed strongest clustering per field for bacterial, Pseudomonas and AOB fingerprints (not shown). No consistent clustering per organic amendment within each field was found among all fingerprints made with the four different primer systems.
Two ecologically distinct parameters, Hill evenness (E) and Shannon diversity (H′), calculated from all fingerprints revealed E values of between 0.514 and 0.784 for bacteria, 0.617 and 0.845 for fungi, 0.623 and 0.848 for Pseudomonas and 0.750 and 0.908 for AOB and H′ values of between 2.875 and 3.501 for bacteria, 3.003 and 3.364 for fungi, 1.837 and 2.826 for Pseudomonas and 2.023 and 2.573 for AOB. No significant differences between average E values were found among organic amendments over the two fields, indicating that there were no interactions between organic amendment and field types. Also, no significant differences were present among values of the same organic amendments averaged over the two fields, indicating that there was no observable effect of the organic amendment type on the evenness of the studied microbial communities. However, E values for bacteria and AOB averaged per field significantly differed between both fields: for bacteria it was 0.657 in field 1 and 0.744 in field 6 and for AOB, 0.797 in field 1 and 0.838 in field 6. E values for fungi and Pseudomonas averaged per field did not differ significantly between both fields. Average Shannon diversity H′ values of the organic amendments over both fields, and per organic amendment of both fields together, never significantly differed from each other. This indicates that diversities of the four studied microbial groups are not influenced by the interaction between organic amendment and field types and also not by organic amendment alone. However, average H′ values for Pseudomonas and AOB per field significantly differed between fields: for Pseudomonas it was 2.295 in field 1 and 2.524 in field 6 and for AOB, 2.199 in field 1 and 2.375 in field 6. No significant effects between average H′ values from both fields were observed for bacteria and fungi. There were, thus, significant effects of field type on the evenness of bacteria, on the diversity of Pseudomonas and on evenness and diversity of AOB.
Effect of organic amendment and field types on R. solani AG2-2IIIB growth in soil
Growth of R. solani AG2-2IIIB in all 32 soils, recorded as post-emergence damping-off in sugar beet plants (blackening of the roots followed by root rot and damping off of the plantlets), was monitored within a period of 25 days. Damping-off in sugar beet plants was not observed in plants grown in the same, but non-inoculated (control) soils within the same time period. R. solani AG2-2IIIB growth rate, measured as the progress in the number of diseased plants over time and expressed as ‘area under the disease progress curve’ (AUDPC) and R. solani AG2-2IIIB growth after 25 days, expressed as cumulative disease incidence (DI), differed per organic amendment and field type.
Average AUDPC and DI values, calculated per organic amendment for each of the two fields, are presented in . There was a statistical interaction between organic amendment and field types on the R. solani AG2-2IIIB growth rate in soil. AUDPC value for CSD amendment was found to be highest in field 1, whereas it was lowest in field 6, when compared with all other organic amendments in both fields. The AUDPC value of CSD amendment in field 1 was statistically different from all other organic amendments in both fields, except for C amendment in field 1 and SD amendment in field 6. There was no statistical interaction between organic amendment and field type on R. solani AG2-2IIIB growth after 25 days. However, after averaging DI values from all organic amendment treatments per field, it revealed that there was a significant effect of field type on R. solani AG2-2IIIB growth after 25 days; i.e. DI values were higher in field 1 (51.7) than in field 6 (39.8). ANOVA performed on arcsin-transformed DI values revealed the same pattern; i.e., no statistical interaction between organic amendment and field type and a significant difference between average values from field 1 and 6. There is thus an interaction between organic amendment and field types on the R. solani AG2-2IIIB growth rate in soils, whereas a measurable effect of field type was found on R. solani AG2-2IIIB growth after 25 days.
Table 2. R. solani AG2-2IIIB growth in soils treated with different organic amendments
Cross comparison between chemical, microbiological and R. solani AG2-2IIIB growth parameters
Biplot diagrams made by redundancy analyses (RDA) on individual bands in the 32 fingerprints from each of the four microbial groups as ‘species’ variables, and on nominal values of organic amendment and field types and on quantitative values of chemical and R. solani AG2-2IIIB growth parameters as ‘environmental’ variables, always revealed a correlation of field type with the first axis for all four microbial groups (). The effect of field type on microbial community structures in soils was significant for bacteria, fungi and AOB (P values of between 0.002 and 0.008) and just above the arbitrary level of significance for Pseudomonas (P = 0.07). Overall, the second axis was correlated with organic amendment types, but the effects of organic amendments on species composition in all four communities were never significant. At three occasions there were effects just above the arbitrary level of significance, i.e. C treatment on the bacterial (P = 0.056), SD treatment on the fungal (P = 0.058) and S treatment on the AOB (P = 0.096) community structures. Generally, bacterial, fungal and AOB communities in field 1 soil segregated further along the second axis than those of field 6 soil, indicating that qualitative effects of organic amendments on these communities were more profound in field 1 soils.
Fig. 1. Biplots calculated by redundancy analysis (RDA) on (a) bacterial, (b) fungal, (c) Pseudomonas and (d) ammonia-oxidizing β-proteobacterial communities, determined by group-specific PCR-DGGEs on DNA extracts from 32 soil samples. Soils were taken from plots located in fields 1 and 6. For RDA, nominal values of organic amendment (S, C, SD, CSD) and field type (1, 6) and quantitative values of C (OM, DOC and Ctot) and total N, NO3-N, pH, and R. solani AG2-2IIIB growth (DI and AUDPC) were used as ‘environmental’ variables. Explanation of symbols: diamonds, field 1; circles, field 6; symbols with diagonal lines, S, grid lines, SD; solid, CSD; open, C. Values at both axes indicates the percentage of the variation explained by each of the axes. Vectors are indicative for the contribution of each of the environmental variables on the community structure; longer sizes and smaller angles with the axis indicate higher correlation with the axis. The level of statistical significance of the interactions with the community structures are indicated by ***, P ≤ 0.01; **, 0.01 > P ≤ 0.05; *, 0.05 > P ≤ 0.1; nothing, P > 0.1.
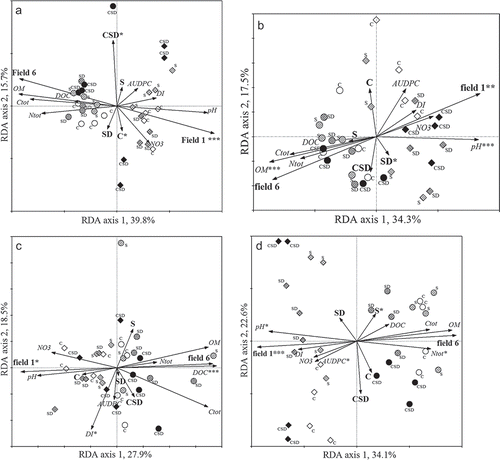
Vectors of the measured chemical parameters pH and OM always pointed into opposite directions and were, at most occasions, longest and showing the smallest angle with the first axis. This indicates that both parameters had an inverse relationship with each other and that both explained highest variation in all four microbial communities. However, significant effects of OM (P = 0.002) and pH (P = 0.008) were only observed on the fungal community structure. One other significant effect was found for DOC on the Pseudomonas community structure (P = 0.008). Effects just above the arbitrary level of significance were only found for total nitrogen content (P = 0.086) and pH (P = 0.088) on the AOB community structure.
Vector lengths of the R. solani AG2-2IIIB growth parameters AUDPC and DI were in general smaller than vectors representing quantitative chemical and nominal field parameters. This indicates that the variation that can be explained by these two parameters is relatively low in comparison with the other parameters. Significant relationships were not found, although at two occasions interactions just above the arbitrary level of significance were found between DI and Pseudomonas (P = 0.056), and between AUDPC and AOB (P = 0.054) communities. This indicates that trends in relationships exist between R. solani AG2-2IIIB growth parameters and the Pseudomonas and AOB species compositions.
Discussion
The combinations of organic amendment with field types, by virtue of differences in organic carbon and nitrogen input into two soils that differed in cropping and agricultural management histories, led to large variations in the measured soil chemical and microbiological parameters and two parameters describing growth of R. solani AG2-2IIIB in soil. No obvious effects of soil organic amendment on all measured parameters were found, with the exceptions of small, but non-significant effects of compost amendment on the bacterial community structure, slurry on the ammonia-oxidizing β-Proteobacteria (AOB) community structure and slurry and dung amendment on the fungal community structure. The effect of field type, however, was evident in three of the four studied communities.
Field type was the factor that was discriminative for most parameters and in field 1 soils, pH and NO3-N were higher and total C, organic matter content, water-dissolvable C, total N, evenness of bacterial and ammonia-oxidizing β-proteobacterial communities and diversities of Pseudomonas and ammonia-oxidizing β-proteobacterial communities were lower than in field 6 soils. Across the board, pH, organic matter content and water-dissolvable organic carbon were the strongest chemical parameters determining the structures of the bacterial, fungal, Pseudomonas and ammonia-oxidizing β-proteobacterial communities in all 32 soils, although their effects were only significant for fungi (pH and organic matter) and Pseudomonas (water-dissolvable organic carbon). How the chemical composition influences the microbial community structure in soils, and how both factors influence pathogen growth in soil are questions that still need to be addressed.
The soils of the two studied fields were extreme with respect to pH and organic matter content. Both fields were located proximate to each other on the same experimental farm and there are no structural or textural differences between the soils at both locations. Also, grass-clover ley and organic management practice were the same for both fields during the 2 years preceding sampling. The chemical and microbiological diversification must have occurred in the period before and shortly after conversion to organic farming. It has been shown before that cropping history is a major driver behind changes in microbial community structures and increased suppression of plant diseases (Garbeva et al., Citation2004, Citation2006; Larkin et al., Citation2010a , 2011). In the studies done by Larkin and coworkers (Citation2010b , 2011), it was demonstrated that cropping histories may have long-lasting effects on disease incidences and crop yields. Also, in the studies done by Garbeva et al. (Citation2004, Citation2006), the effect of crop history was evident and the soil used in these studies was almost identical with that of ours (the location of the experimental site was not further than 500 m from our sites). There, it was shown that damping-off in cauliflower caused by R. solani AG3 was lower in soil with a history of permanent grass coverage than in soil under crop rotation. Also, the Pseudomonas community structure differed between soils with different cropping histories. It can be concluded that the over-2-years difference in cropping histories between our fields was responsible for major differences between chemical and microbiological parameters.
The amendment of compost, slurry and dung to field 1 soil resulted in the highest rate of R. solani AG2-2IIIB growth in soil, whereas this rate was lowest in the same treatment applied to field 6 soil. Opposite effects in R. solani AG2-2IIIB growth thus resulted from the same treatment in different fields. We did not anticipate such a large variation in effects of organic amendments between both fields. This can be a finding of importance for agricultural practices where organic amendments are applied for control of soilborne diseases, as the effect of organic amendment on susceptibility to plant diseases can fluctuate even within the same soil, depending on differences in crop rotations, soil amendments and agricultural management practices.
The structure of the Pseudomonas community was influenced by the size of the water-dissolvable organic carbon fraction in soil. Pseudomonas species generally are considered to be opportunists, rapidly responding to available nutrients in soil (Grünwald et al., Citation2000). Also, individual Pseudomonas species often act as antagonists towards plant pathogens and there was a small, at the level of P = 0.056, relationship between the structure of the Pseudomonas community and cumulative disease incidence after 25 days. Via multivariate analyses, only correlations between measured parameters can be calculated and significance of effects do not necessarily indicate existence of a direct relationship between measured parameters. Still, it is an intriguing observation that lowest R. solani AG2-2IIIB growth was measured in the soil that contained the highest dissolvable organic carbon fraction and the highest diversity of Pseudomonas species (field 6 soil). Most likely, dissolvable organic carbon influences the Pseudomonas community structure in soil by increasing the number of dominant Pseudomonas species. There might be an effect of resident Pseudomonas species on R. solani AG2-2IIIB growth in the studied soils.
One of the groups not expected to be responsive to any variation in carbon availability in soil are the chemolitho-autotrophic ammonium-oxidizing β-proteobacteria. These ammonium-oxidizing β-proteobacteria are chemolithotrophes and do not necessarily need organic substrates for growth. Next to field type, no other significant interactions with any of the measured parameters were found. However, the interaction, just above P = 0.05, with R. solani AG2-2IIIB growth rate in soil was remarkable. To the best of our understanding, there is no direct relationship between ammonium-oxidizing β-proteobacteria and R. solani AG2-2IIIB. Presumably, the same factors affected both the ammonia-oxidizing β-proteobacteria and R. solani AG2-2IIIB in soil. Two of these factors may be total nitrogen content and soil pH, because interactions of these two parameters with ammonium-oxidizing β-proteobacteria were at levels below P = 0.1. The fact that there was also an interaction between the ammonium-oxidizing β-proteobacteria and slurry amendment at the level of P = 0.096 indicates that ammonia present in slurry, or in the total nitrogen pool in soil, can be drivers behind the species composition of this group. Further, it has been shown before that soil pH had an effect on the ammonium-oxidizing β-proteobacterial community structure, mostly comprised of Nitrospira species (Kowalchuk et al., Citation2000). Especially one subgroup of Nitrospira species, denoted as cluster 2 in Kowalchuk et al. (Citation2000), became more dominant in soils with lower pH values. We did not perform further attempts to identify these shifted groups of ammonium-oxidizing β-proteobacteria in our community fingerprints.
The combination of the different agricultural measures applied to our experimental soil plots changed soil chemical and microbiological parameters and it is difficult to assign their causal relationships in soil. Most likely, it is the chemical composition of the soil that drives the microbial community structure; however, it is unknown how the input of micro-organisms associated with the different amendments might change the soil microbial community structure. Further, it may be the soil micro-organisms that interact with the pathogen, but we did not measure effects directly on R. solani AG2-2IIIB mycelium in soil, but indirectly via development of damping-off in sugar beet plants. Decrease in susceptibility for damping-off most likely results from suppression of the pathogen, but changes in the physiology of sugar beet plants by the different circumstances prevailing in the differently treated soils also can play a role. We can conclude, however, that particular chemical parameters like pH, organic matter and water-dissolvable organic carbon contents are responsible for structural changes in communities of particular groups of soil micro-organisms. Growth of R. solani AG2-2IIIB in soil may be influenced by these community shifts. Interactions between particular groups of micro-organisms and R. solani AG2-2IIIB either can be direct, as proposed for the Pseudomonas group of species, or in concert affected by the same factors, which most likely is the case for soil pH and total N on both the ammonium-oxidizing β-proteobacteria and R. solani AG2-2IIIB. To demonstrate direct effects of soil micro-organisms on soilborne pathogens, it is better to focus on interactions between pathogen and antagonizing micro-organisms in soil, or under simulated circumstances representing the soil environment. This ultimately will provide information on the specific microbial groups that are responsible for suppression of phytopathogens and that can report on differences in susceptibility to plant diseases upon application of organic amendments to soil.
Acknowledgements
This research was part of the Ecogenomics programme which is sponsored by the Dutch National Genomics Initiative and the Research programme on sustainable agriculture (KB4) of the Dutch Ministry of Agriculture, Nature and Food Safety. We would like to thank Aad Termorshuizen and Arjen Speksnijder for their assistance in experimental design and helpful discussions, Henny Halm for chemical analyses of the soils, Aikaterini Politikou for disease suppressiveness testing of the soils and workers of the experimental farm ‘De Droevendaal’ for their assistance in field maintenance and soil sampling.
References
- Anderson , I.C. and Cairney , J.W.C. 2004 . Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques . Envir. Microbiol. , 6 : 769 – 779 .
- Ball , D. F. 1964 . Loss-on-ignition as an estimate of organic matter and organic carbon in non-calcareous soils . J. Soil Sci. , 15 : 84 – 92 .
- Doran , J. W. 2011 . Soil microbial and biochemical changes associated with reduced tillage . Soil Sci. Soc. Am. J. , 44 : 765 – 771 .
- Garbeva , P. , Postma , J. , Van Veen , J. A. and Van Elsas , J. D. 2006 . Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3 . Envir. Microbiol. , 8 : 233 – 246 .
- Garbeva , P. , Van Veen , J. A. and Van Elsas , J. D. 2004 . Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness . Ann. Rev. Phytopathol. , 42 : 243 – 270 .
- Gardes , M. and Bruns , T. D. 1993 . ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhiziae and rusts . Mol. Ecol. , 2 : 113 – 118 .
- Gorissen , A. , Van Overbeek , L. S. and Van Elsas , J. D. 2004 . Pig slurry reduces the survival of Ralstonia solanacearum biovar 2 in soil . Can. J. Microbiol. , 50 : 587 – 593 .
- Grünwald , N. J. , Hu , S. and Van Bruggen , A. H. C. 2000 . Short-term cover crop decomposition in organic and conventional soils: characterization of soil C, N, microbial and plant pathogen dynamics . Eur. J. Plant Pathol. , 106 : 37 – 50 .
- Heuer , H. , Krsek , M. , Baker , P. , Smalla , K. and Wellington , E. M. H. 1997 . Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients . Appl. Envir. Microbiol. , 63 : 3233 – 3241 .
- Hoitink , H. A. J. and Boehm , M. J. 1999 . Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon . Annu. Rev. Phytopathol. , 37 : 427 – 446 .
- Janvier , C. , Villeneuve , F. , Alabouvette , C. , Edel-Hermann , V. , Mateille , T. and Steinberg , C. 2007 . Soil health through soil disease suppression: which strategy from descriptors to indicators . Soil Biol. Biochem. , 39 : 1 – 23 .
- Kennedy , A. C. and Smith , K. L. 1995 . Soil microbial diversity and the sustainability of agricultural soils . Plant Soil , 170 : 75 – 86 .
- Kowalchuk , G. A. and Stephen , J. R. 2001 . Ammonia-oxidizing bacteria: a model for microbial ecology . Annu. Rev. Microbiol. , 55 : 485 – 529 .
- Kowalchuk , G. A. , Stephen , J. R. , De Boer , W. , Prosser , J. I. , Embley , T. M. and Woldendorp , J. W. 1997 . Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments . Appl. Envir. Microbiol. , 63 : 1489 – 1497 .
- Kowalchuk , G. A. , Stienstra , A. W. , Heilig , G. H. J. , Stephen , J. R. and Woldendorp , J. W. 2000 . Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands) . FEMS Microbiol. Ecol. , 31 : 207 – 215 .
- Larkin , R. P. , Griffin , T. S. and Wayne Honeycutt , C. 2010a . Rotation and cover crop effects on soilborne potato diseases, tuber yield, and soil microbial communities . Plant Dis. , 94 : 1491 – 1502 .
- Larkin , R. P. , Wayne Honeycutt , C. and Modesto Olanya , O. 2010b . Management of Verticillium wilt of potato with disease-suppressive green manures and as affected by previous cropping history . Plant Dis. , 95 : 568 – 576 .
- Larkin , R. P. , Wayne Honeycutt , C. , Griffin , T. S. , Modesto Olanya , O. , Halloran , J. M. and He , Z. 2011 . Effects of different potato cropping system approaches and water management on soilborne disease and soil microbial communities . Phytopathology , 101 : 58 – 67 .
- Mazzola , M. 2002 . Mechanisms of natural soil suppressiveness to soil borne diseases . Antonie van Leeuwenhoek , 81 : 557 – 564 .
- Muyzer , G. , De Waal , E. C. and Uitterlinden , A. G. 1993 . Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA . Appl. Envir. Microbiol. , 59 : 695 – 700 .
- Pascual , J. A. , Hernandez , T. , Garcia , C. , Lerma , S. and Lynch , J. M. 2002 . Effectiveness of municipal waste compost and its humic fraction in suppressing Pythium ultimum. Microb. Ecol . 44 : 59 – 68 .
- Pérez-Piqueres , A. , Edel-Hermann , V. , Alabouvette , C. and Steinberg , C. 2006 . Response of soil microbial communities to compost amendments . Soil Biol. Biochem. , 38 : 460 – 470 .
- Postma , J. , Scheper , R. W. A. and Schilder , M. T. 2010 . Effect of successive cauliflower plantings and Rhizoctonia solani AG2-1 inoculations on disease suppressiveness of a suppressive and a conducive soil . Soil Biol. Biochem. , 42 : 804 – 812 .
- Postma , J. , Schilder , M. T. , Bloem , J. and Van Leeuwen-Haagsma , W. K. 2008 . Soil suppressiveness and functional diversity of the soil microflora in organic farming systems . Soil Biol. Biochem. , 40 : 2394 – 2406 .
- Salles , J. F. , Van Veen , J. A. and Van Elsas , J. D. 2004 . Multivariate analyses of Burkholderia species in soil: effect of crop and land use history . Appl. Envir. Microbiol. , 70 : 4012 – 4020 .
- Sarniguet , A. , Lucas , P. and Lucas , M. 1992a . Relationships between take-all, soil conduciveness to the disease, populations of fluorescent pseudomonads and nitrogen fertilizers . Plant Soil , 145 : 17 – 27 .
- Sarniguet , A. , Lucas , P. , Lucas , M. and Samson , R. 1992b . Soil conduciveness to take-all of wheat: influence of the nitrogen fertilizers on the structure of populations of fluorescent pseudomonads . Plant Soil , 145 : 29 – 36 .
- Schönfeld , J. , Gelsomino , A. , Van Overbeek , L. S. , Gorissen , A. , Smalla , K. and Van Elsas , J. D. 2003 . Effects of compost addition and simulated solarisation on the fate of Ralstonia solanacearum biovar 2 and indigenous bacteria in soil . FEMS Microb. Ecol. , 43 : 63 – 74 .
- Senechkin , I. V. , Speksnijder , A. G. C. L. , Semenov , A. M. , Van Bruggen , A. H. C. and Van Overbeek , L. S. 2010 . Isolation and partial characterization of bacterial strains on low organic carbon medium from soils fertilized with different organic amendments . Microb. Ecol. , 60 : 829 – 839 .
- Smit , E. , Leeflang , P. , Glandorf , B. , Van Elsas , J. D. and Wernars , K. 1999 . Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis . Appl. Envir. Microbiol. , 65 : 2614 – 2621 .
- Termorshuizen , A. J. , Van Rijn , E. , Van Der Gaag , D. J. , Alabouvette , C. , Chen , Y. , Lagerlöf , J. , Malandrakis , A. A. Paplomatas , E. J. 2006 . Suppressiveness of 18 composts against 7 pathosystems: variability in pathogen response . Soil Biol. Biochem. , 38 : 2461 – 2477 .
- Van Bruggen , A. H. C. and Semenov , A. M. 2000 . In search of biological indicators for soil health and disease suppression . Appl. Soil Ecol. , 15 : 13 – 24 .
- Van Elsas , J. D. and Wolters , A. C. 1995 . “ Polymerase chain reaction (PCR) of soil microbial DNA ” . In Molecular Microbial Ecology Manual , Edited by: AKkermans , A. D. L. , Van Elsas , J. D. and De Bruijn , F.J. Dordrecht, the , , Netherlands : Kluwer Academic Publishers .
- Van Overbeek , L. S. and Van Elsas , J. D. 2008 . Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.) . FEMS Microb. Ecol. , 64 : 283 – 296 .
- White , T. J. T. , Bruns , S. L. and Taylor , J. W. 1990 . “ Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics ” . In PCR Protocols: A Guide to Methods and Applications , Edited by: Innis , M. A. , Gelfand , D. H. , Sninsky , J. J. and White , T. J. New York : Academic Press .
- Widmer , F. , Seidler , R. J. , Gillevet , P. M. , Watrud , L. S. and Di Giovanni , G. D. 1998 . A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples . Appl. Envir. Microbiol. , 64 : 2545 – 2553 .