Abstract
A new fungal disease of inflorescences of Mexican nut (Jatropha curcas L.) was found on plants in two experimental fields located in Sinaloa, Mexico during summer 2011. The fungus caused inflorescence blight on flowers with typical symptoms of dark brown necrotic lesions; both plantations of J. curcas had disease incidence of 50–60%. Based on cultural and morphological characteristics and ribosomal DNA spacer sequences, the pathogen was identified as Alternaria alternata (Fr.) Keissler. To our knowledge, this is the first report of Alternaria inflorescence blight disease in J. curcas in Mexico.
Résumé
Une nouvelle maladie fongique a été observée sur les inflorescences du pignon d'Inde (Jatropha curcas L.) dans deux parcelles expérimentales à Sinaloa, au Mexique, au cours de l'été 2011. Le champignon a causé la brûlure des fleurs et affiché les symptômes typiques, c'est-à-dire des lésions nécrotiques brun foncé. Dans les deux plantations de J. curcas, l'incidence de la maladie était d'environ 50 à 60 %. En se basant sur les caractéristiques culturales et morphologiques ainsi que sur les séquences de l'espaceur de l'ARNr, l'agent pathogène a été identifié en tant qu'Alternaria alternata (Fr.) Keissler. À notre connaissance, il s'agit de la première mention d'une alternariose sur J. curcas au Mexique.
Introduction
The Mexican nut, Jatropha curcas L., an oilseed plant belonging to the Euphorbiaceae family, is increasingly gaining economic importance in tropical and subtropical regions due to its ability to adapt to arid and semi-arid climates as well as its biofuel potential (Heller, Citation1996). However, little is known about agronomic practices, disease problems and management of J. curcas grown in Mexico. In Sinaloa, Mexico, several adaptation trials have been conducted since 2008. In summer 2011, symptoms of flower blight were observed on inflorescences of 3-year-old J. curcas plants of the ecotypes ‘Morelos‘, ‘Puebla’, and ‘Veracruz’ growing in two experimental fields in Sinaloa, Mexico. The symptoms were small dark brown lesions that gradually coalesced to form larger necrotic lesions and finally the flowers dropped. The objective of this study was to identify the causal organism of the inflorescence blight disease of J. curcas.
Materials and methods
Sampling and isolation
Diseased flowers with typical symptoms of dark brown necrotic lesions were collected from two plantations of J. curcas. Samples were cut into small pieces and surface disinfected with 0.6% sodium hypochlorite solution for 1 min, rinsed thoroughly with sterile distilled water, the moisture was removed with sterilized filter paper, and the pieces were cultured on potato dextrose agar (PDA) plates and incubated in the dark at 28 °C. Microscopic observations were conducted by mounting fungal tissues of 10-day-old colonies in water and morphological characteristics of conidia were described.
Pathogenicity test
Pathogenicity of the fungus was confirmed twice (two experiments) by inoculating healthy inflorescences of J. curcas plants grown in an experimental field. Ten inflorescences per experiment were sprayed with ∼5 mL of conidial suspension (1 × 106 conidia mL−1) obtained from 15-day-old culture plates. Before inoculation, all inflorescences, inoculated and water-sprayed controls, were disinfected with 0.1% sodium hypochlorite solution for 1 min and rinsed thoroughly with sterile distilled water. After inoculations, control and inoculated inflorescences were covered with polythene bags for 8 days.
Molecular identification
DNA extracted from four isolates and seven re-isolates of inoculated inflorescences was amplified by Polymerase Chain Reaction (PCR) with ITS1 (5´-TCCGTAGGTGAACCTGCGG-3´) and ITS4 (5´-TCCTCCGCTTATTGATATGC-3´) primers resulting in fragments of ∼550 bp (White et al., Citation1990). PCR products of four isolates and four re-isolates were sequenced in both directions and searched for homology to those of other species. On the basis of the ITS distances of one isolate and a re-isolate sequences (JQ660836 and JQ660841, respectively) and of sequences obtained from GenBank database, a similarity dendrogram was constructed for the ITS2 region using clustal W alignment method with weighed residue weight table in the program MegAlign of the sequence analysis software suite Lasergene (DNASTAR Inc.) (Konstantinova et al., Citation2002).
Results and discussion
Inflorescence blight disease affected both plantations of Jatropha curcas in Sinaloa, Mexico. The observed symptoms were small dark brown lesions that gradually coalesced to form larger necrotic lesions, and finally caused flowers to drop. The same fungus was consistently isolated on PDA from all infected samples and no other fungi were isolated from the infected flowers. The fungus had dark-coloured mycelium (), conidia were obclavate, obpyriform, ovoid or ellipsoidal, in chains, dark, large, with 1–7 transverse and 1–3 longitudinal septa, 13.97–57.5 × 6.35–20.32 m (). The isolated fungus was identified as Alternaria alternata based on cultural and morphological characteristics (Simmons, Citation2007).
Fig. 1. Morphology of Alternaria alternata and inflorescences of Jatropha curcas. a, Mycelial growth of fungal pathogen on PDA. b, Conidial mass of A. alternata at 40×. c, Inoculated inflorescence with Alternaria alternata after 8 days of the inoculation. d, Control inflorescence.
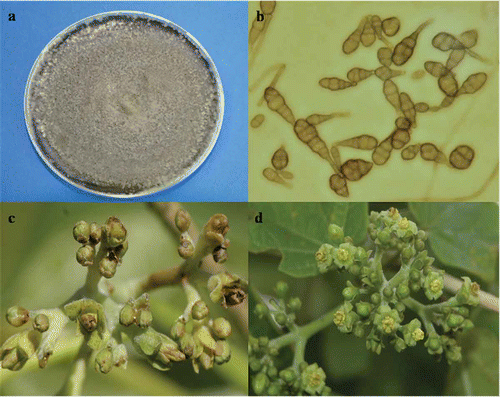
Pathogenicity tests conducted on healthy inflorescences of J. curcas resulted in the development of typical symptoms of the disease on flowers after 6–8 days (), whereas no symptoms were observed on control inflorescences (). The same pathogen was consistently re-isolated and identified from all inoculated inflorescences, confirming Koch's postulates.
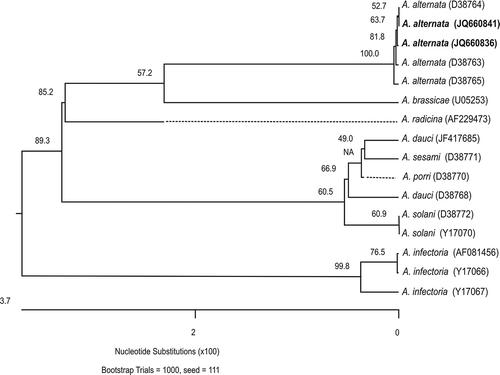
The ITS sequences (GenBank accession numbers JQ660836 to JQ660843) showed 100% of identity not only among all isolates from J. curcas but also to other Alternaria alternata isolates deposited in GenBank. The tree of ITS2 region showed that all isolates of A. alternata clustered together in a monophyletic group with 100% similarity (). In contrast, A. dauci, A. sesame, A. porri and A. solani clustered in a monophyletic group, correlating with the fact that these species produce larger-size conidia (Simmons, Citation2007).
Fig. 2. Similarity dendrogram on distances derived from sequences of the ITS2 regions of Alternaria species. Distances were determined by clustal W alignment method with weighed residue weight table in the program MegAlign of the sequence analysis software suite Lasergene (DNASTAR Inc.). Bootstrap (confidence) values are shown on the branches. Alternaria alternata strains from this study are in bold.
In this study, the incidence of inflorescence blight caused by A. alternata was 50–60% in the two plantations of J. curcas, which significantly reduced fruit production. Alternaria spp. cause common diseases on leaves, stems, fruits and flowers of a wide range of plant species (Agrios, Citation2005). Flower blight incited by A. alternata on several hosts has been reported previously. These hosts include chickpea (Cicer arietinum) (Vishwakarma & Chaudhary, Citation1974), wax flower (Chamelaucium uncinatum) (Taylor et al., Citation1998), desert zinnia (Zinnia acerosa) (Colbaugh et al., Citation2001), and himalayan incarvillea (Incarvillea emodi) (Shanmugam et al., Citation2011). To our knowledge, this is the first report of A. alternata causing inflorescence blight on Jatropha curcas in Mexico.
Acknowledgements
We thank Consejo Nacional de Ciencia y Tecnología (CONACYT) Project I0110/127/11, C-197-11 and Instituto Politécnico Nacional for financial support of this research project.
References
- Agrios , G.N. 2005 . Plant Pathology , Fifth , 922 San Diego , CA : Elsevier Academic Press . edition
- Colbaugh , P.F. , Mackay , W.A. and George , S. 2001 . Alternaria alternata flower blight of Zinnia acerosa in Texas . Plant Dis , 85 : 228
- Heller , J. 1996 . “ Physic nut. Jatropha curcas L ” . In Promoting the conservation and use of underutilized and neglected crops , Gatersleben : Institute of Plant Genetics and Crop Plant Research/Rome: International Plant Genetic Resources Institute .
- Konstantinova , P. , Bonants , P.J.M. , van Gent-Pelzer , M.P.E. , van der Zouwen , P. and van den Bulk , R. 2002 . Development of specific primers for detection and identification of Alternaria spp. in carrot material by PCR and comparison with blotter and plating assays . Mycol. Res , 106 : 23 – 33 .
- Shanmugam , V. , Dhyani , D. and Ananthapadmanaban , D. 2011 . First report of Alternaria sp. causing blight on Incarvillea emodi . Australas. Plant Dis. Notes , 6 : 33 – 35 .
- Simmons , E.G. 2007 . Alternaria. An Identification Manual. (CBS Biodiversity Series 6) , Utrecht , , the Netherlands : CBS Fungal Biodiversity Centre .
- Taylor , M.N. , Wearing , A.H. , Joyce , D.C. and Simons , D.H. 1998 . Alternaria alternata causes petal blight and flower drop in harvested Geraldton waxflower . Australas. Plant. Pathol , 3 : 207 – 210 .
- Vishwakarma , S.N. and Chaudhary , K.C.B. 1974 . Floral blight of gram incited by Alternaria alternata . Neth. J. Plant Pathol , 80 : 110 – 112 .
- White , T.J. , Buns , T. , Lee , S. and Taylor , J. 1990 . “ Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics ” . In PCR Protocols , Edited by: Innis , M. , Gelfand , D.H. , Sninsky , J.J. and White , T.J. 315 – 322 . San Diego , CA : Academic Press .