Abstract
Seventy-eight mungbean genotypes were grown in the field and 28 genotypes were found to be resistant to Mungbean yellow mosaic virus, while the remainder were susceptible. All genotypes were further subjected to agroinoculation to confirm their resistance/susceptibility against MYMV using two different MYMV strains, namely VA 221 and VA 239. The results showed that only three genotypes, namely ML 1108, KMG 189 and SP 84, exhibited resistance to the VA 221 strain while 77 genotypes were found to be susceptible to strain VA 239. Only one genotype, namely ML 818, showed resistance to strain VA 239, but it was susceptible to strain VA 221. The accumulation of the viral genome in the host was confirmed by PCR amplification of the coat protein gene of MYMV using gene-specific primers.
Résumé
Soixante-dix-huit génotypes de haricot mungo ont été cultivés en champ et 28 se sont avérés résistants au virus de la mosaïque jaune du haricot mungo (VMJA), tandis que les autres y étaient réceptifs. Afin de confirmer leur résistance ou leur susceptibilité au VMJA, tous les génotypes ont de plus été soumis à l'agro-inoculation avec deux souches différentes de VMJA, notamment les souches VA 221 et VA 239. Les résultats ont montré que seulement 3 génotypes, ML 1108, KMG 189 et SP 84, ont affiché de la résistance à l’égard de la souche VA 221, tandis que 77 génotypes se sont avérés réceptifs à la souche VA 239. Un seul génotype, ML 818, a affiché de la résistance à l’égard de la souche VA 239, mais il était réceptif à la souche VA 221. L'accumulation du génome viral chez l'hôte a été confirmée par amplification par PCR du gène de la protéine de coque du VMJA à l'aide d'amorces spécifiques.
Introduction
Mungbean [Vigna radiata (L.) Wilczek] is one of the most important pulse crops grown in Asia, particularly in India. It is known for its digestability, good flavour and high protein content. However, yellow mosaic disease (YMD), caused by Mungbean yellow mosaic virus (MYMV), is one of the most devastating diseases that limits mungbean production throughout Asia, including India. Usharani et al. (Citation2004) reported that MYMV is more predominant in the southern regions of India where it causes severe yield loss in farmer's fields in Tamil Nadu province. If the plants are infected at the seedling stage, it can lead to 85–100% loss in yield.
Diverse virus isolates (genetically distinct strains, reassortants and recombinants) of MYMV may have different levels of stability or virulence, as reflected by the symptom severity in each genotype of mungbean. These isolates of MYMV have been classified into two distinct strains, and reassortants between the two strains have been identified based on nucleotide sequencing (Balaji et al., Citation2004). The development of resistant cultivars is the most effective and economical strategy against MYMV (Karthikeyan et al., Citation2012). However, current screening procedures rely upon natural field infection. In the absence of a uniform inoculation technique, reliable results on cultivar response cannot be obtained. MYMV is not transmitted mechanically but is transmitted by the whitefly vector, Bemisia tabaci, in a persistent (circulative) manner. The efficacy of transmission and behaviour of whiteflies varies with the host genotype, virus strains and growth conditions. Hence, plant breeders and pathologists are in need of a biological/molecular tool that can lead to the identification of MYMV resistant/susceptible genotypes.
In this context, the advances in the field of genetic engineering have shown that agroinoculation is a useful method to screen for virus-resistant plants in disease-resistance breeding (Biswas & Varma, Citation2001; Bi et al., Citation2010; Karthikeyan et al., Citation2011). Previous asynchronous/failed infections have improved since the use of agroinoculation (Vaghchhipawala et al., Citation2011). Expression of both DNA/RNA and mono/bipartite virus genomes using infectious viral clones can be achieved using Agrobacterium tumefaciens which serves as an alternative route for viral infection of plants by using the Ti plasmid (Grimsley et al., Citation1986). As a result, a genome-size copy of viral DNA is released; it replicates, is encapsidated, systemically spreads and expresses disease symptoms. Previous researchers working on urdbean, mungbean and soybean (Jacob et al., Citation2003; Usharani et al., Citation2005; Haq et al., Citation2010; Karthikeyan et al., Citation2011) demonstrated the feasibility of using an in vitro agroinoculation procedure in MYMV studies.
The objective of this research was to identify mungbean resistant genotypes against two MYMV strains based on field screening and agroinoculation, to evaluate its usefulness in mungbean breeding for MYMV resistance.
Materials and methods
Plant materials
In this study, we used 78 mungbean genotypes, originating from different parts of India. Seeds of these genotypes were provided by the National Pulse Research Centre, Pudhukottai in Tamil Nadu, India and are used for several mungbean breeding programmes.
Screening of mungbean genotypes against MYMV
The seeds of 78 mungbean genotypes were sown in three replications during two consecutive seasons, namely July to October, 2006 and March to June, 2007 at the National Pulse Research Centre. The plants were maintained properly by providing row to row and plant to plant spacing at 50 cm and 10 cm, respectively. The infector row method, in which two test rows alternating with spreader rows of the susceptible cultivar C05 were sown, was adopted in the field to evaluate MYMV infection. Summer whiteflies are the source of the virus in the field, so no insecticide was sprayed in order to maintain the natural whitefly population in the experimental field. When 80% of the plants showed MYMV symptoms, scoring of the test materials was done. The rating scale suggested by Singh et al. (Citation1988) was adopted. The mean disease score was calculated on the basis of disease rating and frequency of diseased plants per total number of plants. Based upon the MYMY score, the mungbean plants were divided into two groups – resistant and susceptible. Plants that are resistant (R) and moderately resistant (MR) were included in the resistant group, while moderately susceptible (MS), susceptible (S) and highly susceptible (HS) plants were categorized in the susceptible group.
MYMV isolate and strain details
Balaji et al. (Citation2004) previously collected MYMV infected leaves from urdbean fields in Vamban, Pudukkottai district in Tamil Nadu, India. They cloned the MYMV genome from the infected tissues and constructed the infectious clones, namely VA 221 (KA30 DNA A + KA22 DNA B) and VA 239 (KA30 DNA A + KA27 DNA B). In addition, the infectious clones were mobilized in strains of A. tumefaciens, namely Ach 5 and C 58. The two mobilized strains of MYMV (VA 221 and VA 239) were obtained from this research laboratory (Balaji et al., Citation2004) and were used for this study to screen the mungbean germplasm.
Agroinoculation
Agroinoculation study was conducted in the Centre for Plant Molecular Biology, Tamil Nadu Agricultural University, during the period 2007–2008. Agroinoculation was done on surface-sterilized overnight sprouted seeds of the 78 mungbean genotypes according to the protocol of Jacob et al. (Citation2003). Agroinoculated plants were maintained in a growth chamber at 25 °C, 60–70% relative humidity and a photoperiod of 16 h light/18 h dark. Hoagland's solution was applied twice a week for proper growth of the plants and symptom development was recorded from the 7th day after inoculation in the trifoliate leaves. The presence of yellow mosaic symptoms at a given time was scored as susceptible and the absence was scored as resistance against the disease. The percentage infection in the mungbean plants through agroinoculation was calculated based on the number of plants infected to the ratio of the number of plants inoculated.
DNA extraction and PCR analysis
Genomic DNA was isolated from 78 agroinoculated mungbean genotypes following the modified protocol of Karuppanapandian et al. (Citation2006). The quality and quantity of DNA were checked by agarose gel electrophoresis. The final concentration of all the samples was adjusted to 25 ng μL−1. The DNA templates from all the agroinoculated mungbean accessions were amplified using the MYMV coat protein (CP) gene-specific primers that were designed based on the CP gene sequences of MYMV isolates from Tamil Nadu that were deposited in the NCBI database. PCR amplification was performed in a PTC Thermal Cycler 100 (MJ Research Inc., San Francisco, CA). A volume of 15 μL of PCR mix contained 1.5 μL of 10× assay buffer, 0.5 μL of 2.5 mM dNTPs (Bangalore Genei Ltd., India), 0.20 μL of 0.5 unit μL−1 of Taq polymerase (Bangalore Genei Ltd.), 1.0 μL of 10 μM primer (First Base, Singapore) and 3.0 μL of 25 ng μL−1 DNA. The temperature cycles were as follows: 5 min at 94 °C followed by 35 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C. The final elongation step was extended to 5 min at 72 °C and finally maintained at 4 °C. The amplified products were separated on a 1.5% agarose gel.
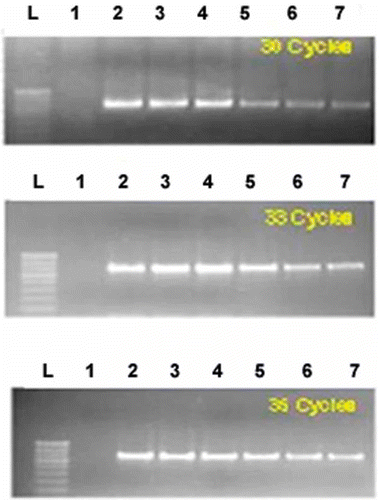
Quantification of virus in the agroinoculated plants
To quantify the viral DNA concentration in the agroinoculated plants, DNA was extracted from leaf samples collected at different days (5, 10 and 15) after inoculation along with uninoculated controls. PCR conditions were optimized at 30, 33 and 35 cycles to check the difference in the viral load. The integrated density value (IDV) was calculated using the analysis tool in the alpha imager for every band with the standard values from the ladder (100 bp). A standard curve (cubic spiline) was drawn against standard sample in 250 ng and the IDV.
Results and discussion
The available evidence gathered since the 1960s suggests that the diversity of crop plants and the geographical area affected by the incidence of MYMV has increased gradually. The agriculturally significant hosts of MYMV include mungbean, urdbean (Vigna mungo), soybean (Glycine max), cowpea (Vigna unguiculata) and common bean (Phaseolus vulgaris) (Malathi & John, 2008). In breeding programmes, resistance to MYMV was previously determined by visual symptomatology. Symptomless genotypes were assumed to be resistant; however, since lines can be infected without showing symptoms, it is possible that they are not resistant. In addition, MYMV symptoms do not always appear in the field, making it difficult to identify resistant lines. In this regard, artificial inoculation is a useful screening strategy to identify virus-resistant lines and to obtain a more uniform disease epidemic compared with natural infestation. Maintaining the natural inocula of the virus isolates is a challenge since MYMV is not transmitted mechanically but is transmitted by the whitefly vector, Bemisia tabaci, in a persistent (circulative) manner. Therefore, constructs of the virus isolates inoculated into plants via agroinoculation were used to evaluate resistance of the genotypes at the molecular level. We obtained results of screening in the field followed by screening with agroinoculation against MYMV in 78 mungbean germplasm lines. Infection by MYMV was evaluated using the rating scale suggested by Singh et al. (Citation1988). According to the mean disease score, the mungbean genotypes were categorized into five groups – resistant (R, 15 genotypes), moderately resistant (MR, 13 genotypes), moderately susceptible (MS, 12 genotypes), susceptible (S, 13 genotypes) and highly susceptible (HS, 25 genotypes) (). We used urdbean MYMV isolates VA 221 (KA30 DNA A + KA22 DNA B) and VA 239 (KA30 DNA A + KA27 DNA B) for agro-inoculation screening. The isolates from urdbean produce two types of yellow mosaic symptoms in mungbean (Balaji et al., Citation2004) and are more virulent compared with other MYMV isolates. In addition these isolates are very common in Tamil Nadu province in India.
Table 1. Infectivity responses of mungbean genotypes to agroinoculation
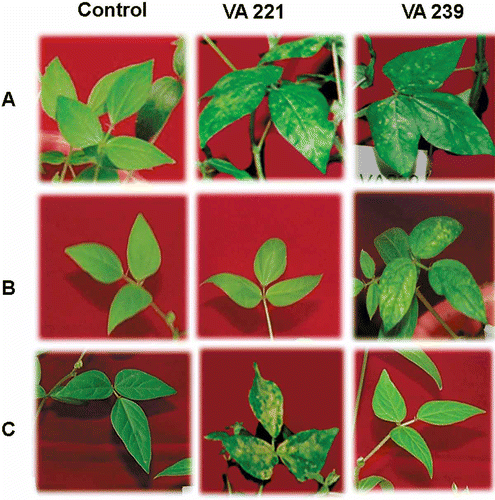
All the 78 mungbean genotypes (50 field-susceptible genotypes and 28 field-resistant genotypes) were subjected to agroinoculation in two replications. The susceptible genotypes were included in agroinoculation studies to standardize the agroinoculation protocol. Among the 78 genotypes, all 50 field susceptible genotypes exhibited typical yellow mosaic symptoms in the trifoliate leaves upon agroinoculation, using both Agrobacterium strains. Milder symptoms of yellow mosaic were observed 2 weeks after inoculation in the trifoliate leaves of plants inoculated with VA221 (KA30 DNA A and KA22 DNA B) strain. The strain VA 239 (KA 30 DNA A and KA27 DNAB) had very severe effects on the morphology of plants in addition to the development of typical yellow mosaic symptoms. The plants were stunted with shortened petioles, trifoliate leaves curling downwards and were sterile (). Interestingly, among the 28 field-resistant genotypes, three genotypes, namely ML1108, KMG189 and SP84, did not develop mosaic or leaf curling symptoms upon inoculation with VA221 strain () but they exhibited susceptibility against the other strain VA239. One entry, namely ML 818, showed resistance against the strain VA239 and it was found to be susceptible to the strain VA221. To confirm the results, the experiment (28 field-resistant genotypes alone) was repeated twice and comparable results were obtained. The above results clearly indicated that most of the “field-resistant” lines were susceptible. The resistance exhibited at the field level may be due to some mechanisms which prevent the entry of the virus through insect vectors. The level of infectivity of these agroinoculated plants ranged between 0–100%. Three representative genotypes from each category (R, MR, MS, S and HS) are shown in Usharani et al. (Citation2005) performed infectivity analysis of a soybean isolate of Mungbean yellow mosaic virus by agroinoculation and obtained similar results, i.e. about 71–95% MYMV infectivity which is similar to the present study. Interestingly, the field-resistant genotypes also showed 100% infection in the agroinoculation studies (data not shown). Under field conditions, natural infection may not produce accurate results even if the studies are conducted in fields with high vector populations.
Fig. 1. A) ML-558 (Field susceptible genotype) reaction to VA 221 and VA 239 strains. B) SP 84 (Field resistant genotype) reaction to VA 221 and susceptible reaction to VA 239. C) ML 818 (Field resistant genotype) susceptible reaction to VA 221 and resistant reaction to VA 239.
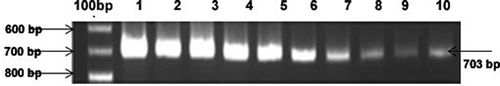
The agroinoculated mungbean plants started developing yellow mosaic symptoms from the 13th day to the 25th day and there were no symptoms in the uninoculated control plants. At the 25th day, when the yellow mosaic symptoms were clearly seen on the leaves, the leaf samples were collected for DNA isolation. The PCR confirmatory studies were done to verify the presence of viral DNA in the host genome using oligonucleotide primers that are specific to the MYMV coat protein gene of DNA A (the expected amplicon size being 703 bp) in all infected samples. These results are in accordance with the reports of Usharani et al. (Citation2005) indicating the presence of viral DNA in agroinoculated symptomatic plants and their absence in asymptomatic plants. The previous MYMV screening results reported by Karthikeyan et al. (Citation2011) did not detail the mechanism of resistance and the accumulation of virus in the agroinoculated plants. Understanding the resistant mechanism and quantifying the viral load in the plants are very helpful to evaluate the virus resistance in the plants. To determine the accumulation of MYMV genome (VA 221) in the host mungbean genome, PCR amplification of MYMV coat protein gene was carried out using different concentrations of genomic DNA (0.3125 ng to 50 ng) isolated from the agroinoculated plants of mungbean. The intensity of PCR product decreased as the concentration of DNA decreased ().
Fig. 2. PCR confirmatory analysis of viral geneome as revealed by concentration dependent PCR reaction (field resistant mungbean ML1108). Lane 1 : 50 ng, Lane 2 : 40 ng, Lane 3 : 30 ng, Lane 4 : 20 ng, Lane 5 : 10 ng ; Lane 6 : 5 ng, Lane 7 : 2.5ng, Lane 8 : 1.25ng, Lane 9 : 0.625ng, Lane 10 : 0.1325ng.
Understanding the mechanism behind MYMV resistance in an asymptomatic line
The DNA from leaf samples collected at 5, 10 and 15 days after inoculation were used to quantify the viral DNA load in the agroinoculated genotype ML 818. When optimized at 30, 33 and 35 cycles of PCR, for difference in the viral load, there was a comparable trend observed between the two strains (). The intensity of the band was the same for all the cycles for VA221 but a decreasing intensity was observed for VA239 for every corresponding decrease from 35 to 33 up to 30 cycles. Besides, the integrated density value (IDV) was calculated and a standard curve (cubic spiline) was drawn against a standard sample in 250 ng and the IDV. The standard curve for 30 cycles depicted a higher IDV value for VA221 on all the three days of sampling and was lower in VA239. The standard curve for 33 cycles depicts the higher IDV values for VA221 (at 5, 10 and 15 days). IDV was higher (5 days) and lower (10 and 15 days) in strain VA239 .The standard curve for 35 cycles showed almost higher values for both strains. It is clear that lesser viral load can be detected in resistant plants when the number of cycles are decreased to have low copy numbers. In most of the published methods, the number of PCR cycles required to detect viruses in symptomatic plants was relatively low (25), but in some instances, including asymptomatic plants, the more sensitive, real-time PCR assay was able to detect virus where conventional PCR assays did not (Yadav et al., Citation2008). These reports suggested that the conventional PCR assays can easily detect virus where the viral load is high and are sensitive enough to detect virus where the viral load is low. As per Jones et al. (Citation2008) a generic PCR detection method for tobraviruses could detect viruses of higher viral load but failed in case of lesser viral load. The new identified germplasm (resistant sources) can be used in the mungbean breeding programme for improving resistance to MYMV.
Acknowledgements
We thank the Department of Bio-Technology, a division under the Department of Science and Technology, India, for funding and supporting the research programme. We are grateful to Dr K. Veluthambi, Madurai Kamaraj University, India, for providing the Agrobacterium strains for our work.
References
- Balaji , V. , Vanitharani , R. , Karthikeyan , A.S. , Anbalagan , S. and Veluthambi , K. 2004 . Infectivity analysis of two variable DNA B components of Mungbean yellow mosaic virus-Vigna in Vigna mungo and Vigna radiata . J. Biosci. , 29 : 297 – 308 .
- Bi , H. , Aileni , M. and Zhang , P. 2010 . Evaluation of cassava varieties for cassava mosaic disease resistance jointly by agroinoculation screening and molecular markers . Afr. J. Plant Sci. , 4 : 330 – 338 .
- Biswas , K. and Varma , A. 2001 . Agroinoculation: a method of screening germplasm resistance to mungbean yellow mosaic geminivirus . Indian Phytopathol. , 54 : 240 – 245 .
- Grimsley , N. , Hohn , B. , Hohn , T. and Walden , R. 1986 . Agroinfection, an alternative route for viral infection of plants by using the Ti plasmid . Proc. Natl. Acad. Sci. USA , 83 : 3282 – 3286 .
- Haq , Q.M. , Arif , A. and Malathi , V.G. 2010 . Engineering resistance against Mungbean yellow mosaic India virus using antisense RNA . Indian J. Virol. , 21 : 82 – 85 .
- Jacob , S.S. , Vanitharani , R. , Karthikeyan , A.S , Chinchore , Y. , Thillaichidambaram , P. and Veluthambi , K. 2003 . Mungbean yellow mosaic virus-Vi agroinfection by co delivery of DNA A and DNA B from one Agrobacterium strain . Plant Dis. , 87 : 247 – 251 .
- Jones , D. , Farreyrol , K. , Clover , G.R.G. and Pearson , M.N. 2008 . Development of generic PCR detection method for tobraviruses . Aust. Plant Pathol. , 37 : 132 – 136 .
- Karthikeyan , A. , Sudha , M. , Pandiyan , M. , Senthil , N. , Shobhana , V.G. and Nagarajan , P. 2011 . Screening of MYMV resistant mungbean (Vigna radiata (L) Wilczek) progenies through agroinoculation . Int. J. Plant Pathol. , 2 : 115 – 125 .
- Karthikeyan , A. , Sudha , M. , Senthil , N. , Pandiyan , M. , Raveendran , M. and Nagarajan , P. 2012 . Screening and identification of RAPD markers linked to MYMV resistance in mungbean (Vigna radiata (L) Wilczek) . Arch. Phytopathol. , 45 : 712 – 716 .
- Karuppanapandian , T. , Karuppudurai , T. , Sinha , A.P. , Hamarul Haniya , M. and Manoharan , K. 2006 . Genetic diversity in green gram [Vigna radiata (L.)] landraces analyzed by using random amplified polymorphic DNA (RAPD) . Afr. J.Biotech. , 5 : 1214 – 1219 .
- Malathi , V.G. and John , P. 2008 . “ Geminiviruses infecting legumes ” . In Characterization, Diagnosis and Management of Plant Viruses: Vegetables and Pulses Crops , Edited by: Rao , G.P. , Lava Kumar , P. and Holguin-Pena , R.J. 97 – 123 . Houston , TX : Stadium Press LLC .
- Singh , G. , Kapoor , S. and Singh , K. 1988 . “ Multiple disease resistance in mungbean with special emphasis on Mungbean yellow mosaic virus ” . In Proceedings of the Second International Symposium on Mungbean Vegetable Research and Development Centre Edited by: Shanmugasundaram , S. 290 – 296 . Tainan , , Taiwan
- Usharani , K.S. , Surendranath , B. , Haq , Q.M.R. and Malathi , V.G . 2004 . Yellow mosaic virus infecting soybean in northern India is distinct from the species infecting soybean in southern and western . India Curr. Sci. , 86 : 845 – 850 .
- Usharani , K.S. , Surendranath , B. , Haq , Q.M.R. and Malathi , V.G. 2005 . Infectivity analysis of a soybean isolate of Mungbean yellow mosaic India virus by agroinoculation . J. Gen. Plant Pathol. , 71 : 230 – 237 .
- Vaghchhipawala , Z. , Clemencia , M. , Senthilkumar , M. and Kirankumar , S. 2011 . Agroinoculation and agroinfiltration: simple tools for complex gene function analyses . Methods Mol. Biol. , 678 : 65 – 76 .
- Yadav , N.R. , Pankaj , B. , Supriya , M.R. and Yadav , R.C. 2008 . “ Engineering disease resistance in crop plants ” . In Advances in Plant Biotechnology , Edited by: Rao , G.P. , Yipeng , Z. , Radchuk , V.V. and Bhatnagar , S.K. 527 – 539 . Houston , TX : Stadium Press LLC .