Abstract
Coconut palms from the Grand-Lahou region of Côte d’Ivoire exhibiting lethal yellowing-like symptoms and infected by a phytoplasma were further tested for the characterization of the phytoplasma pathogen based on the 16S rRNA gene analysis. Nested PCR with phytoplasma universal and lethal yellowing-specific primers resulted in amplification of products of expected sizes from seven out of 17 symptomatic trees tested, while non-infected symptomless plants yielded no DNA amplification. The 16S rDNA sequence of the phytoplasma detected from Grand-Lahou in coconut trees (KC999037) affected with the Côte d’Ivoire Lethal Yellowing disease showed a 99% sequence identity with that of the Ghanaian lethal yellowing phytoplasma (Cape St. Paul Wilt strain) of group 16SrXXII. The 16S rDNA sequence-based virtual and actual RFLP and phylogenetic analyses revealed that the phytoplasma associated with Côte d’Ivoire Lethal Yellowing in Grand Lahou is as a member of a new subgroup within group 16SrXXII that was designated as 16SrXXII-B. The present results support previous suspicions of disease spread to Côte d’Ivoire from neighbouring Ghana, posing a threat to the survival of the coconut trees in the Grand-Lahou region and for the Ivorian coconut industry.
Résumé
Les cocotiers de la région de Grand-Lahou en Côte d’Ivoire présentant les symptômes du jaunissement mortel et infectés par un phytoplasme ont été davantage testés pour caractériser l’agent pathogène en se basant sur l’analyse du gène d’ARNr 16S. La PCR par amorces incluses avec amorces universelles de phytoplasmes et amorces spécifiques du jaunissement mortel a entraîné l’amplification de produits de longueurs attendues chez sept des 17 arbres symptomatiques testés, tandis que les plants sains et ne présentant aucun symptôme n’ont pas amplifié l’ADN. La séquence de l’ADNr 16S du phytoplasme détecté chez les cocotiers (KC999037) de Grand-Lahou souffrant de jaunissement mortel en Côte d’Ivoire, a affiché un taux de similarité de 99% avec celle du phytoplasme du groupe16SrXXII (souche de la flétrissure de Cap St. Paul) responsable du jaunissement mortel au Ghana, pays voisin. Les analyses virtuelles et réelles du RFLP basé sur la séquence de l’ADNr 16S et les analyses phylogénétiques ont révélé que le phytoplasme associé au jaunissement mortel de Grand-Lahou en Côte d’Ivoire est un membre d’un nouveau sous-groupe issu du groupe16SrXXII, désigné 16SrXXII-B. Les résultats actuels appuient les soupçons antérieurs concernant la provenance ghanéenne de la maladie qui s’est propagée dans la Côte d’Ivoire, ce qui constitue une menace à la survie des cocotiers de la région de Grand-Lahou et pour l’industrie ivoirienne de la noix de coco.
Introduction
Coconut (Cocos nucifera L.) is considered the most important crop along the coastal belt of West Africa which provides food and income for many women and landless poor with minimum capital outlay. Côte d’Ivoire is among the first 20 out of 92 world coconut-producing countries (UNCTAD Citation2012), and is the top African, Caribbean & Pacific exporter of coconut oil (from copra), that nowadays accounts for 2.5% of the world vegetable oil production. In Côte d’Ivoire (Allou et al. Citation2012), coconut palm is cultivated on approximately 50 000 hectares (1 to 5 ha/farmer), and produces an average of 45 000 tons of copra/year, the main source of income for people living in the coastal region.
Lethal yellowing (LY)-type phytoplasma diseases have become the major threat for the global coconut industry (Nipah et al. Citation2007) in Africa and the Caribbean basin, and affect C. nucifera and 35 other species of palms in the Americas. Symptoms are variable among palm genera and, in the case of coconuts, among cultivars (Harrison et al. Citation2008). It is the pattern of appearance and chronological progression of symptoms that accurately identifies the disease.
Despite the similarities in symptoms of LY with coconut lethal decline (LD) diseases that occur in Africa (Sullivan & Harrison Citation2013), there are a number of epidemiological considerations, such as geographical distribution patterns, rates of spread, and varietal and host species susceptibility that have indicated dissimilarities among phytoplasmas and vector species involved with these diseases. To acknowledge these differences, they are collectively referred to as lethal yellowing (LY)-like diseases.
LY-like diseases occurring in West Africa include ‘maladie de Kaincopé’ in Togo, ‘Awka wilt’ or ‘lethal decline’ (LD) in Nigeria, ‘Kribi’ in Cameroon, and ‘Cape St. Paul Wilt Disease’ (CSPWD) in Ghana. In East Africa, LY or ‘lethal decline’ (LD) has been reported in Tanzania (LDT), Mozambique (LYM), Benin, Equatorial Guinea (Bonnot et al. Citation2010) and Kenya.
Devastating losses of coconut plantations have been associated with such LY-type phytoplasma diseases. In Tanzania, ‘lethal decline’ annihilated about 56% of the southern palms in the last 30 years (Mpunami et al. Citation1999). In Nigeria, ‘Akwa wilt’ killed over 98% of the West African Tall (WAT) palms in 11 years (Odewale et al. Citation2010), and CSPWD has destroyed about one million coconut trees of the Ghanaian coconut production in the last 30 years (Nipah et al. Citation2007).
Phytoplasmas are bacteria-like plant pathogens that are transmitted by phloem-feeding insects and cause devastating yield losses in diverse crops worldwide (Strauss, Citation2009; Duduk & Bertaccini Citation2011). They have been studied using DNA-based methods due to their failure to grow in artificial culture media. Differentiation and classification of phytoplasmas therefore rely on molecular analyses of conserved genes, in particular the 16S rRNA. Several hundred phytoplasma strains have been classified on the basis of distinct 16S rRNA gene RFLP patterns resolved on actual and/or virtual electrophoresis gel analysis (Lee et al. Citation1998; IRPCM Citation2004; Wei et al. Citation2007).
LY phytoplasmas occurring in the Caribbean belong to group 16SrIV ‘Coconut Lethal Yellowing’. However, in this group, the number of subgroups has been continuously increasing in recent years, now reaching six subgroups (Harrison et al. Citation2008; Dollet et al. Citation2011). Until the end of the 1990s, phytoplasmas associated with ‘maladie de Kaincopé’ in Togo, ‘Awka wilt’ in Nigeria and CSPWD in Ghana were listed within the 16SrIV group (Lee et al. Citation1998; Harrison et al. Citation2008). Recently, they were included in a new group, 16SrXXII (Wei et al. Citation2007), incidentally cited as ‘Candidatus Phytoplasma cocosnigeriae’, where particularly the phytoplasma associated with ‘Awka wilt’ was classified as a new subgroup designated 16SrXXII-A (Tymon et al. Citation1998; IRPCM Citation2004).
Over the past 10 years, a LY-like disease named as Côte d’Ivoire Lethal Yellowing (CILY) has been spreading through the Ivorian coastal coconut plantations, destroying more than 350 ha and causing losses of about 12 000 tons of copra/year (ProMED Citation2013). CILY symptoms include leaf yellowing starting in the older leaves and quickly moving to the young ones, drying of spikelets progressing to blackening of the whole inflorescence, immature fruit drop, rotting of heart tissues, and crown death of the palm within six months of initial symptom appearance, leaving a scene of bare trunks, typically known as ‘telephone poles’. These symptoms resembled those caused by the phytoplasma associated with ‘Cape St. Paul Wilt Disease’ in Ghana. The cause of CILY had not been found until Canadian and Ivorian scientists recently identified the phytoplasma associated with the CILY outbreak of the Grand-Lahou region (Konan Konan et al. Citation2013) as an isolate closely related to the CSPWD phytoplasma strain of Ghana.
CILY has become a phytosanitary risk for the Ivorian coconut industry, and particularly for the Ivorian multi-site International Coconut Gene Bank that provides service for Africa and the Indian Ocean region. CILY phytoplasma is now threatening more than 7000 hectares of coconut palms in the Grand-Lahou Department of Côte d’Ivoire in the Lagunes Region (ProMED Citation2013). The present study was undertaken to characterize the phytoplasma pathogen associated with the CILY outbreak in the Grand-Lahou region to support the further development of strategies to more effectively monitor and control the disease. This involved PCR, sequencing and RFLP analyses of the 16Sr RNA gene of the CILY phytoplasma, and included comparisons with phytoplasmas associated with LY-like and LD-like diseases from the Caribbean and West and East Africa to establish their phylogenetic relationships, and provide tools to determine the possible origin of the pathogen spread.
Materials and methods
Coconut plant material and total DNA extraction
A total of 17 coconut trees of ‘West African Tall’ (WAT) variety and ‘PB121’ (‘Malayan Yellow Dwarf’ × WAT) coconut hybrids showing CILY symptoms were surveyed for the CILY phytoplasma at randomly selected plots of the Grand-Lahou. Samples originated from coconut trees showing symptoms of nut drop, necrosis of immature inflorescences, progressive frond yellowing with brown older leaves (), and eventual death of palms showing bare trunks, known as ‘telephone poles’ (). Tissue samples from leaves, stem apexes, hearts and inflorescences of CILY symptomatic (17 samples) and symptomless (eight samples) coconut palms were collected during January–March 2013, and 100 mg of tissue was subjected to a small-scale total DNA extraction (N’nan Citation2004). Total DNA of a phytoplasma from group 16SrI ‘Candidatus Phytoplasma asteris’ extracted from infected periwinkle (Catharantus roseus L.), kindly provided by Dr A. Bertaccini (University of Bologna, Italy), was used as a positive control for PCR assays.
Fig. 1 Symptoms of lethal yellowing associated with the CILY phytoplasma in the Grand-Lahou region. (A) Coconut palms showing frond yellowing progressing to the younger leaves; older leaves are brown and will eventually dry up and die. (B) Coconut palms showing bare trunks corresponding to the last stage of the disease, known as ‘telephone poles’.

Polymerase chain reaction
Total DNA was used as a template for nested PCR assays with universal primers that target the phytoplasma 16S rRNA gene. Primers included R16mF2/R1 (Gundersen & Lee Citation1996) for the first PCR reaction, and R16F2n/R2 for the nested reaction (Gundersen & Lee Citation1996). Primers G813F/GAKSR (Tymon et al. Citation1998) were used for the specific detection of the CSPWD phytoplasma strain. For all PCR reactions, 50 ng of DNA template was added to a 25 µL PCR reaction (GE Healthcare, UK). For the nested reaction, 1 µL of the first-round PCR product was used. Thirty-five cycles were performed for all primer pairs. PCR cycling conditions for R16mF2/R1 and R16F2n/R2 primer pairs were as follows: 1 min (2 min for the initial denaturation) at 94 ºC, 2 min at 50 ºC and 3 min (8 min for the final extension) at 72 ºC. For primers G813F/GAKSR, the annealing temperature was 53 ºC. Five microlitres of the PCR products were separated in a 1.5% agarose gel and visualized with SYBR Safe (Invitrogen, USA) in a UV gel documenter (Alpha Innotech, USA).
Cloning, sequencing and sequence analysis
Representative R16F2n/R2 PCR amplicons were purified on spin columns (Omega Bio-Tek, USA), cloned (pGEM-T Easy Vector, Promega), and sequenced bi-directionally. The consensus 16S rDNA sequence was compared by BLAST (Altschul et al. Citation1990) with reference sequences in GenBank, including representative phytoplasmas of 16SrIV and 16SrXXII groups. Sequences obtained were aligned using Clustal W (Thompson et al. Citation1994) and phylogenetic trees were constructed using the neighbour-joining method with the program MEGA version 4.0 (Kumar et al. Citation2004) with default values and 1000 replicates for bootstrap analysis.
In silico and actual restriction fragment length polymorphism (RFLP)
The trimmed and aligned R16F2n/R2 sequence of the CILY phytoplasma and those of representative phytoplasmas of groups 16SrIV and 16SrXXII were exported to the in silico restriction analysis and virtual gel plotting program pDRAW32, developed by AcaClone software (http://www.acaclone.com). Each aligned DNA fragment was digested in silico with AluI, BfaI, BstUI (ThaI), HaeIII, HinfI, HpaI, HpaII, MseI, RsaI, TaqI and Tsp509I restriction endonucleases. After in silico restriction digestion, a virtual 3.0% agarose gel electrophoresis image with minimum 50 bp was plotted automatically to the computer screen for RFLP profiles comparison and determination of signature banding.
The virtual RFLP patterns were compared and a similarity coefficient (F) was calculated for each pair of phytoplasma strains (Lee et al. Citation1998) according to the formula F = 2Nxy/(Nx + Ny), in which Nx and Ny are the total number of bands resulting from digestions by the 11 restriction endonucleases in strains × and y, respectively, and Nxy is the number of bands shared by the two strains. Ten microlitres of R16F2n/R2 purified PCR products of the CILY phytoplasma were digested with representative restriction endonucleases (following manufacturer’s recommendation) to support in silico RFLP results. RFLP profiles were visualized in a 3% agarose gel stained with SYBR Safe (Invitrogen, USA) in an UV gel documenter (Alpha Innotech, USA).
Results
Detection of phytoplasma DNA in coconut palms
Phytoplasma DNA was amplified from seven out of 17 CILY samples from symptomatic trees. No PCR amplicons were obtained from the symptomless tissue samples. CILY phytoplasma detection was consistent and reproducible for the affected coconut palms when DNA was derived from either spear leaves, inflorescences or heart tissues. Failure of PCR amplification from the 10 remaining symptom-bearing samples may be attributed to the inability to sample the pathogen in mature plant tissues (Harrison et al. Citation1994) due to the irregular distribution of phytoplasma rather than its absence from these tissues.
In silico and actual restriction fragment length polymorphism (RFLP)
Restriction endonucleases yielded unique AluI, BfaI, HaeIII, HpaI, HpaII, MseI, TaqI and Tsp509I RFLP patterns for the CILY phytoplasma that clearly differentiated it from those phytoplasma strains of groups 16SrIV and 16SrXXII (). Similar coefficients derived from virtual RFLP analysis of the R16F2n/R2 sequence of the CILY phytoplasma were compared with those of selected phytoplasma strains from groups 16SrIV and 16SrXXII (). CILY phytoplasma scored values less than the threshold (0.97) to delineate a new RFLP subgroup (Wei et al. Citation2007). Similarity coefficient values less than 0.97 also range among 16SrIV strains that conform distinct subgroups like 16SrIV-A, 16SrIV-B, 16SrIV-C and 16SrIV-D. Based on the RFLP profiles and the similarity coefficient values, the CILY phytoplasma may be designated as a new subgroup within the group 16SrXXII. AluI, HaeIII, HpaI, HpaII and MseI actual RFLP profiles () corresponded with the in silico RFLP patterns, supporting the identification of the CILY phytoplasma as a member of group 16SrXXII, and a new subgroup 16SrXXII-B.
Fig. 2 Virtual RFLP generated from in silico digestion of the R16F2n/R2 sequences of the CILY phytoplasma and representatives from 16SrIV and 16SrXXII phytoplasma groups with AluI, BfaI, HaeIII, HpaI, HpaII and MseI. MW: phiX174DNA-HaeIII digest DNA molecular weight marker. Lane 1: CILY(Cote d’Ivoire)-16SrXXII-B (KC999037); Lane 2: CSPWD(Ghana)-16SrXXII (JQ868442); Lane 3: CSPWD(Ghana)-16SrXXII (Y13912); Lane 4: LDN(Nigeria)-16SrXXII-A (Y14175); Lane 5: LDT(Tanzania)-16SrXXII-C (FJ217385); Lane 6: LYM(Mozambique)-16SrXXII (EU549768); Lane 7: LDY(Yucatan)-16SrIV-B (U18753); Lane 8: LYJ(Jamaica)-16SrIV-A (JX560537); Lane 9: TPPD(Florida)-16SrIVD (AF434989).
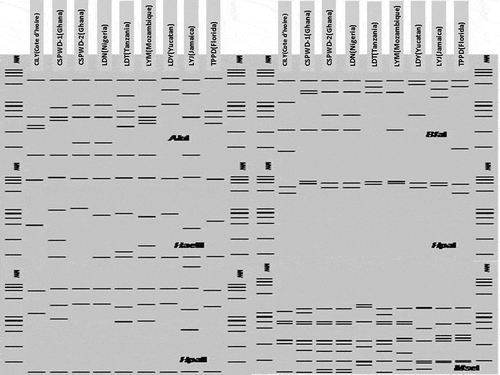
Fig. 3 AluI, HaeIII, HpaI, HpaII and MseI actual RFLP patterns of the CILY phytoplasma visualized in a 3% agarose gel. Lane 1: 100 bp molecular weight marker (MM) (BioLabs, USA). Lanes 2, 3, 4, 5 and 6: AluI, HaeIII, HpaI, HpaII and MseI RFLP profiles.
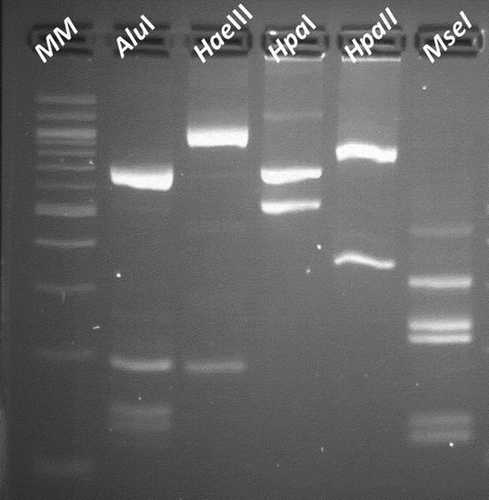
Table 1. Reference strains of ‘Ca. Phytoplasma’ species and GenBank accession numbers used for RFLP and phylogenetic analyses.
Sequence and phylogenetic analysis
The BLAST analysis showed that the R16F2n/R16R2 sequence of the CILY phytoplasma (KC999037) exhibited a highest sequence identity of 99.7% with that of the CSPWD phytoplasma strain from Ghana (JQ868442). Phylogenetic analysis () based on the 16S rDNA sequence supported results from the in silico RFLP () and the sequence () analyses. Phytoplasma sequences used in the phylogeny analysis are shown in . The CILY phytoplasma (KC999037), identified in coconut trees in the Grand-Lahou region, clustered in the same phylogenetic branch that included phytoplasmas associated with coconut and palm diseases in Africa, Central America and the Caribbean that belong to groups 16SrIV and 16SrXXII. The CILY phytoplasma was closely related to the 16SrXXII group since it clustered in the same sub-branch that embraces the 16SrXXII phytoplasmas: CSPWD strains from Ghana (JQ868442 and Y13912), the LY phytoplasma from Mozambique (EU549768), and the LD phytoplasma from Nigeria (Y14175) that belongs to subgroup 16SrXXII-A.
Fig. 4 Phylogenetic tree constructed using the neighbour-joining algorithm based on the R16F2n/R2 sequences of the CILY phytoplasma and reference 16Sr phytoplasma groups. ‘Ca. P. sp’: ‘Candidatus Phytoplasma sp.’ A. laidlawii: Acholeplasma laidlawii is used as the outgroup to root the tree. Phytoplasma sequences are described in .
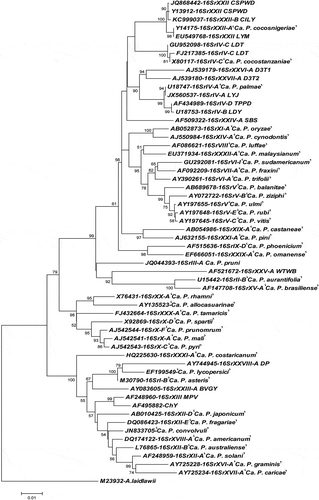
Table 2. Similarity coefficients derived from RFLPs based on putative restriction-site analysis of nucleotide 16S rRNA gene sequences of the CILY phytoplasma (16SrXXII-B) and representative phytoplasmas of 16SrXXII and 16SrIV subgroups.
Table 3. Sequence similarities based on the 16S rDNA of the CILY phytoplasma compared with those representatives of 16SrIV and 16SrXXII phytoplasma subgroups.
Phytoplasma members of 16SrIV subgroups: 16SrIV-A, 16SrIV-B, 16SrIV-D, separated from subgroup 16SrIV-C indistinctly in a single phylogenetic branch. Sequence identities shared by phytoplasmas within the above-mentioned subgroups ranged from 93.9% to 99.9%. This supports the fact that 16SrXXII phytoplasmas differentiate as a single cluster from those embracing the 16SrV subgroups -A, -B, -D and -C, with 16S rDNA sequence identities that ranged from 92.8% to 99.9% ().
Discussion
CILY phytoplasma was infrequently detected from leaves, hearts and inflorescences, which suggests that it is not particularly ubiquitous in the Ivorian coconut palms. This is consistent with previous reports regarding the detection of African LY phytoplasmas in coconut (Mpunami et al. Citation1999). The nested PCR technique has been used for diagnostic purposes particularly in trees and insect vectors when phytoplasmas occur in low titre in the phloem vessels of their host plants (Nejat & Vadamalai Citation2010). Our results confirm the robustness of nested PCR using universal phytoplasma primers to detect and identify phytoplasma strains present in different tissues of phytoplasma-infected coconut palms, and with low titre in the symptomatic host (Nipah et al. Citation2007; Yankey et al. Citation2009).
In Côte d’Ivoire, earlier reports refer to the presence of phytoplasmas in coconut trees affected with Blast, the main nursery disease of oil palm in Africa (Konan-Konan et al. Citation2008). However, no previous official records of a LY-type disease in coconut palms, or its association with a phytoplasma, were available until an isolate of the Ghanaian CSPWD phytoplasma was recently identified from coconut palms showing CILY symptoms (Konan Konan et al. Citation2013). The findings prompted joint efforts to characterize the phytoplasma strain that is currently affecting the coconut trees of the coastal region of the Grand-Lahou area.
RFLP typing of PCR-amplified 16S rRNA gene sequences has become the best approach to classify phytoplasmas into 16Sr groups and subgroups of strains (Duduk & Bertaccini Citation2011). This approach has been also used to classify those lethal yellowing (LY)-type phytoplasmas of group 16SrIV ‘Coconut Lethal Yellows’ in different subgroups (Sullivan & Harrison Citation2013). The subgroup 16SrIV-A causes palm and coconut lethal yellowing in the USA, Central America and the Caribbean (Harrison et al. Citation1994, Citation2002, Citation2008). The subgroup 16SrIV-B is associated with the Yucatan coconut lethal decline (Harrison et al. Citation2002) and coyol palm decline in Honduras (Roca et al. Citation2006).
In Kenya and Tanzania, the subgroup 16SrIV-C, incidentally cited as ‘Candidatus Phytoplasma cocostanzaniae’ is responsible for the coconut lethal disease (IRPCM, Citation2004). The subgroup 16Sr-D causes palm decline diseases in Mexico (Cordova et al. Citation2000) and USA (Ntushelo et al. Citation2013), while 16SrIV-E is associated with coconut lethal decline in Dominican Republic (Martinez et al. Citation2008) and Jamaica (Brown & Mclaughlin Citation2011), and 16SrIV-F is associated with Washingtonia robusta decline in Florida, USA (Harrison et al. Citation2008).
Based on the unique actual and virtual RFLP patterns obtained after in silico enzymatic digestion of the R16F2n/R2 16S rDNA sequence (KC999037) and the coefficient similarity values (), the CILY phytoplasma detected in Ivorian coconut palms was clearly confirmed as a member of a new subgroup within the group 16SrXXII, termed 16SrXXII-B. The virtual RFLP analysis proved a suitable approach for the identification and differentiation of the CILY phytoplasma from the previously described 16SrIV group and 16SrXXII subgroup and has provided information to extend the known western-most distribution of lethal yellowing phytoplasma in West Africa.
The fact that the CILY phytoplasma identified in the Ivorian Grand-Lahou region in 7/17 trees of the ‘West African Tall’ palms and ‘PB121’ coconut hybrids is an isolate of the Ghanaian strain CSPWD, suggests that the CILY disease may be quickly spreading among the susceptible coconut ecotypes. This also highlights the complex epidemiology of two very closely related phytoplasmas that affect the same host in two different and nearby geographic locations. Although the possible spread routes need to be investigated, results support previous suspicions of CILY phytoplasma spreading from neighbouring Ghana, and colonizing the most susceptible local coconut varieties and hybrids in Grand-Lahou.
CILY phytoplasma possesses a great threat for the survival of the coconut germplasm at the ‘Marc Delorme’ Research Station, the main international supplier of coconut germplasm during the past 20 years located just 120 km from the Grand-Lahou CILY outbreak. It requires an urgent assessment of the phytosanitary situation of the new 16SrXXII-B subgroup in the coconut production areas of the Grand-Lahou region towards identifying potential sources of CILY resistance, as well as new effective management strategies to prevent disease spread.
Acknowledgements
We acknowledge the Côte d’Ivoire FIRCA (Fonds Interprofessionnel pour la Recherche et le Conseil Agricoles)/Cocotier CNRA – COC No. 588 that provided funds to carry out this research.
References
- Acaclone pDRAW32 SOFTWARE. Available from http://www.acaclone.com.
- Altschul S, Gish W, Miller W, Meyers E, Lipman D 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Allou K, Issali AE, Lekadou T, Konan-Konan JL, Zakra N, Kouassi P, Bourdeix R, Morin JP, Saraka YDM 2012. Comparative synergetic effect of coconut palm (Cocos nucifera L.) slices and bunches residue of oil palm (Elaeis guineensis JACQ.) associated with two kinds of pheromone traps on Oryctes monoceros OLIVIER trapping in Côte d’Ivoire. Int J Emerg Tech Adv Eng. 2:1–6.
- Bonnot F, de Franqueville H, Lourenço E 2010. Spatial and spatiotemporal pattern analysis of coconut lethal yellowing in Mozambique. Phytopathology. 100:300–312. doi:10.1094/PHYTO-100-4-0300
- Brown SE, McLaughlin WA 2011. Identification of lethal yellowing group (16SrIV) of phytoplasmas in the weeds Stachytarpheta jamaicensis, Macroptilium lathyroides and Cleome rutidosperma in Jamaica. Phytopath Mollicutes. 1:27-34. doi:10.5958/j.2249-4669.1.1.004
- Cordova I, Oropeza C, Almeyda H, Harrison NA 2000. First report of a phytoplasma associated leaf yellowing syndrome of Palma Jipi plants in southern México. Plant Dis. 84:807. doi:10.1094/PDIS.2000.84.7.807A
- Dollet M, Macome F, Vaz A, Fabre S 2011. Phytoplasmas identical to coconut lethal yellowing phytoplasmas from Zambesia (Mozambique) found in a pantomide bug in Cabo Delgado province. Bull Insectol. 64:139–140.
- Duduk B, Bertaccini A 2011. Phytoplasma classification: taxonomy based on 16S ribosomal gene, is it enough?. Phytopath Mollicutes. 1:3–13. doi:10.5958/j.2249-4669.1.1.001
- Gundersen DE, Lee IM 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Medit. 35:144–151.
- Harrison NN, Helmick EE, Elliot M 2008. Lethal yellowing-type diseases of palms associated with phytoplasmas newly identified in Florida, USA. Ann Appl Biol. 153:85–94. doi:10.1111/j.1744-7348.2008.00240.x
- Harrison NA, Myrie W, Jones P, Carpio ML, Castillo M, Doyle MM, Oropeza C 2002. 16S rRNA interoperon sequence heterogeneity distinguishes strain populations of palm lethal yellowing phytoplasma in the Caribbean region. Ann Appl Biol. 141:183–193. doi:10.1111/j.1744-7348.2002.tb00211.x
- Harrison NA, Richardson PA, Kramer JB, Tsai JH 1994. Detection of the mycoplasma-like organism associated with lethal yellowing disease of palms in Florida by polymerase chain reaction. Plant Pathol. 43:998–1008. doi:10.1111/j.1365-3059.1994.tb01649.x
- IRPCM. Phytoplasma/Spiroplasma Working Team–Phytoplasma Taxonomy Group 2004. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol. 54:1243–1255. doi:10.1099/ijs.0.02854-0
- Konan Konan JL, Allou K., Atta Diallo H., Saraka Yao D., Koua B., Kouassi N., Benabid R., Michelutti R., Scott J., Arocha-Rosete Y. 2013. First report on the molecular identification of the phytoplasma associated with a lethal yellowing-type disease of coconut palms in Côte d’Ivoire. New Dis. Rep. 28:3–. [http://dx.doi.org/10.5197/j.2044-0588.2013.028.003]
- Konan Konan JL, Bourdeix R, George ML 2008. Regeneration guidelines: coconut. In: Dulloo ME, Thormann I, Jorge MA, Hanson J, editors. Crop specific regeneration guidelines [CD-ROM]. Rome, Italy: CGIAR System-wide Genetic Resource Programme. (pp. 10).
- Kumar S, Tamura K, Nei M 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150–163. doi:10.1093/bib/5.2.150
- Lee IM, Gundersen-Rindal DE, Davis RE, Bartoszyk IM 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. Int J Syst Evol Microbiol. 48:1153–1169.
- Martinez RT, Narvaez M, Fabre S, Harrison NA, Oropeza C, Dollet M, Hichez E 2008. Coconut lethal yellowing on the southern coast of the Dominican Republic is associated with a new 16SrIV group phytoplasma. Plant Pathol. 57:366. doi:10.1111/j.1365-3059.2007.01726.x
- Mpunami AA, Tymon A, Jones P, Dickinson MJ 1999. Genetic diversity in the coconut lethal yellowing disease phytoplasmas of East Africa. Plant Pathol. 48:109–114. doi:10.1046/j.1365-3059.1999.00314.x
- N’nan O. 2004. Utilisation des biotechnologies pour les échanges et la conservation des ressources génétiques du cocotier (Cocos nucifera L.) [dissertation]. France: Université d’Angers.
- Nejat N, Vadamalai G 2010. Phytoplasma detection in coconut palm and other tropical crops. Plant Pathol J. 9:112–121. doi:10.3923/ppj.2010.112.121
- Nipah JO, Jones P, Dickinson MJ 2007. Detection of lethal yellowing phytoplasma in embryos from coconut palms infected with Cape St Paul Wilt Disease in Ghana. Plant Pathol. 56:777–784. doi:10.1111/j.1365-3059.2007.01623.x
- Ntushelo K, Elliott MM, Harrison N 2013. Palm yellows phytoplasmas and their genetic classification. Afr J Biotech. 12:3376–3382.
- Odewale JO, Odionwaya G, Osagie JI, Ahanon JM. 2010. Rate of lethal yellowing disease (LYD) spread in coconut (Cocos nucifera L) plantation of tall interplanted Dwarf varieties. Proceedings of the 19th Conference of Botanical Society of Nigeria, Umaru Musa Yar”addua University, Katsina, Nigeria. (pp. 47).
- ProMED-mail. 2013. Undiagnosed disease, coconut palm, Côte d’Ivoire: (Lagunes). Int Soc Infect Dis. [Internet]. [revised 2013 Feb 22; cited 2013 Nov 14]. Available from: http://www.promedmail.org]
- Roca MM, Castillo MG, Harrison NA, Oropeza C 2006. First report of a 16SrIV group phytoplasma associated with declining coyol palms in Honduras. Plant Dis. 90:526. doi:10.1094/PD-90-0526B
- Strauss E 2009. Phytoplasma research begins to bloom. Science. 325:388–390. doi:10.1126/science.325_388
- Sullivan M, Harrison N. 2013. CPHST Pest Datasheet for ‘Candidatus Phytoplasma palmae’ and related strains. [Internet]. [revised 2013 June; cited 2013 Nov 14]. USDA-APHIS-PPQ-CPHST. Available from http://caps.ceris.purdue.edu/.
- Thompson JD, Higgins DG, Gibson TJ 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 22:4673–4680. doi:10.1093/nar/22.22.4673
- Tymon AM, Jones P, Harrison NA 1998. Phylogenetic relationships of coconut phytoplasmas and the development of specific oligonucleotide PCR primers. Ann Appl Biol. 132:437–452. doi:10.1111/j.1744-7348.1998.tb05220.x
- UNCTAD, United Nations Conference on Trade and Development. 2012. INFOCOMM– Commodity Profile Coconut. [Internet]. [revised 2012 Nov 12; cited 2013 Nov 14]. http://www.unctad.info/en/Infocomm/AACP-Products/COMMODITY-PROFILE---Coconut2/.
- Wei W, Davis RE, Lee IM, Zhao Y 2007. Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. Int J Syst Evol Microbiol. 57:1855–1867. doi:10.1099/ijs.0.65000-0
- Yankey EN, Pilet F, Quaicoe RN, Dery SK, Dollet M, Dzogbefia VP 2009. Search for alternate hosts of the coconut Cape Saint Paul Wilt Disease pathogen. Ol Corps Gras Li. 16:123–126. doi: 10.1051/ocl.2009.0250
