Abstract
Allele specific (AS) SNP primers were developed for rapid detection of Fusarium oxysporum f.sp. vasinfectum (FOV) race 3. These primers were designed based on single nucleotide polymorphisms of partial sequence alignment of the β-tubulin (BT) gene from several FOV races. The primers showed high specificity for FOV race 3, and exclusively amplified PCR products from FOV race 3 DNAs and total genomic DNAs of FOV race 3-infected plant tissues. These primers did not produce any amplification products for races 1, 2, 4, 6, 7 and 8 of FOV or other FOV and Fusarium species from infected plant tissues. The assay is useful for specific and rapid detection of Fusarium oxysporum f. sp. vasinfectum race 3.
Résumé
Les amorces SNP spécifiques de l’allèle ont été développées en vue de la détection rapide de la race 3 de Fusarium oxysporum f. sp. vasinfectum (FOV). Ces amorces ont été conçues à partir du polymorphisme mononucléotidique de l’alignement de séquences partielles du gène de la tubuline ß de plusieurs races de FOV. Les amorces ont affiché un haut degré de spécificité quant à la race 3 de FOV et aux produits exclusivement amplifiés par PCR de l’ADN de la race 3 et de tout l’ADN génomique des tissus provenant de plantes infectées par la race 3 de FOV. Ces amorces n’ont engendré aucun produit résultant de l’amplification des races 1, 2, 4, 6, 7 et 8 de FOV ni d’autres espèces de FOV et de Fusarium provenant de plantes infectées. Le biotest s’avère utile pour la détection rapide et précise de la race 3 de Fusarium oxysporum f. sp. vasinfectum.
Introduction
Fusarium oxysporum Schlect. f.sp. vasinfectum (FOV) (Atk.) Snyd & Hans is an important asexual species and is well represented among the soilborne fungi all over the world (Burgess Citation1981). F. oxysporum includes morphologically indistinguishable pathogenic, nonpathogenic, and even beneficial strains. The pathogenic strains cause diseases such as vascular wilt, yellowing, root rot, and damping-off in a wide range of economically important crops (Beckman & Roberts Citation1995), while the nonpathogenic strains are defined as strains for which no specific host plants are infected (Lievens et al. Citation2008).
Conventionally, the identification of fungal pathogens has been based solely on morphological characteristics. However, for a number of genera of fungi, the precise definition of species, especially the identification of subspecies (forma speciales or genotypes called races), involves certain difficulties associated with overlapping morphological features and time-consuming mycological analyses.
To date, rapid and accurate methods of Fusarium species and subspecies identification have been developed (Assigbetse et al. Citation1994; Fernandez et al. Citation1994; Attitalla et al. Citation2004; Bogale et al. Citation2006; Bayraktar et al. Citation2008). Abd-Elsalam et al. (Citation2003), using ITS region sequences, designed two primer pairs ITS-Fu-f and ITS-Fu-r which could differentiate F. oxysporum f. sp. vasinfectum, F. moniliforme and F. solani. Moreover, on the basis of 16S and 23S ribosomal subunits of rDNA, Abd-Elsalam et al. (Citation2006) developed specific primers for detection of F. oxysporum f. sp. vasinfectum. Polymorphisms of ribosomal DNA (rDNA) were successfully used by Zambounis et al. (Citation2007) to identify Australian isolates of F. oxysporum f.sp. vasinfectum . They developed specific SNP primers for detection of rDNA intergenic region (IGS) polymorphisms which were able to identify new formae speciales of F. oxysporum in Australia by using PCR-RFLP and Real-Time PCR methods. Along with rDNA, there are a number of single copy genes widely used for identification of formae speciales and races of F. oxysporum.
A large collection of sequences of single copy genes of the Fusarium genus has already been published and deposited in the NCBI, GenBank (Lievens et al. Citation2008). The first sequence-specific primers developed for the translational elongation factor (TEF-1) and mitochondrial small subunit ribosomal DNA (mtSSU) were able to identify many species in the Fusarium genus (Baayen et al. Citation2000, Citation2001; O’Donnell et al. Citation2000; Skovgaard et al. Citation2001). Later, the nitrate reductase (NIR), phosphate permease (PHO) (Skovgaard et al. Citation2001), UTP-ammonia ligase (UTP-ammonia ligase), and trihotretsena 3-acetyltransferase (trichothecene 3-oacetyltransferase (O’Donnell et al. Citation2000) genes also were used for taxonomic and ecotype identification of Fusarium species. However, to the best of our knowledge, molecular approaches are limited to rapidly and specifically detect individual FOV races.
In Uzbekistan, local ecotypes of F. oxysporum f.sp. vasinfectum are poorly studied. For years, the dominant pathogen in cotton was verticillium wilt; however, over the last 10 years, fusarium wilt became a widespread pathogen in most cotton growing areas of the country. For many decades in Uzbekistan, fusarium wilt was known to affect primarily Pima cotton varieties, and long staple Pima-derived Upland cotton cultivars (Marupov et al. Citation2010). Reports from 2007 to 2013 demonstrated that several Upland cotton varieties also became highly susceptible to Fusarium in Uzbekistan. On some farms, yield loss to fusarium wilt reached 85% (Abdullaev et al. Citation2011; Marupov et al. Citation2012; Egamberdiev et al. Citation2013). Recent studies have also shown the presence of races 1, 2, 3, 6 and 7 in Uzbekistan (Egamberdiev et al. Citation2013). Much attention has been paid to FOV race 3, which is wide-spread in the northern part of the country. Large northern cotton growing areas of Uzbekistan, such as Syrdarya and Tashkent, are mainly affected with FOV race 3. The main problem of rapid race identification of FOV races is associated with time to perform procedures and expensive DNA sequencing methodology. The objective of present study was to develop a new and highly specific molecular approach for detecting FOV race 3, using single base extension methods and allele specific PCR (AS-PCR) based on single nucleotide polymorphism (SNP).
Material and methods
Fungal isolates and DNA extraction
F. oxysporum f. sp. vasinfectum isolates used in this study included FOV 316 (race 3), FOV 347 (race 3) (Egamberdiev et al. Citation2013) and other known races that served as controls (races 1, 2, 4, 6, 7, 8) (Kim et al. Citation2005; Ulloa et al. Citation2013). These fungal isolates were obtained from the Plant Pathogens Collection of the Institute of Genetics and Plant Experimental Biology, Tashkent, Uzbekistan and originally isolated from infested soils and diseased cotton plants in Uzbekistan. Isolates FOV 316 and FOV 347 were fully characterized in a previous study by partial sequences of translational elongation factor (EF-1α), ribosomal DNA (rDNA), and β-tubulin (BT) genes (Egamberdiev et al. Citation2013). Total DNA was extracted according to the protocol of Nazar et al. (Citation1991).
Nucleotide sequence analysis
Beta-tubulin (BT) gene-specific primers BT-3 (5’CGTCTAGAGGTACCCATACCGGCA; and BT-5 (5’GCTCTAGACTGCTTTCTGGCAGACC) reported by Tooley et al. (Citation2001) were used to amplify target regions of selected FOV isolates. Fragments were amplified in 25 µL reaction mixture containing 20 ng of genomic DNA, 2.5 µL of 10 × PCR buffer (10 mM Tris-HCl, pH 8.3; 1.5 mM MgCl2; 50 mM KCl), 0.1 mg/mL BSA, 200 µM each of dNTP, 50 pmol of each primer and 1 U of Taq DNA polymerase (Applied Biosystems, Foster City, CA). The PCR reactions were run in a thermocycler Gene Amp PCR System 9700 (Applied Biosystem Foster City, CA). The PCR profile was 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 90 s, with a final extension at 72 °C for 5 min for BT3-BT5. Negative controls (master-mix without DNA template) were included in each experiment. Amplification products were separated on a 1.5% agarose gel and visualized by AlphaImager gel documentation system (v. 5.5, Alpha Innotech Corporation, San Leandro, CA) after staining with ethidium bromide. The products were purified with polyethylene glycol (PEG) solution (containing 26% PEG 8000, 6.5 mM MgCl2 and 0.6 mM sodium acetate pH 5.2) to remove remaining primers. Purified BT fragments were sequenced in both directions using the same primers (BT3 and BT5) and BigDye terminator sequencing kit v. 3.1 on Genetic Analyzer 3130xl (Applied Biosystems, Foster City, CA).
Primer design
Sequencher 4.9 software (Gene Codes Corp. USA) was used for aligning sequences to detect single nucleotide polymorphisms in partial sequence of BT gene. Primers were designed based on sequence information either downstream or upstream from target nucleotide. Identified race-specific SNP were used for design of allele-specific PCR (ASPCR) and SNaPshot primers. Forward primer for single base extension SNP detection was designed to anneal right before SNP. In the case of ASPCR, a primer with race-specific nucleotide on 3’ end was designed to anneal strictly on SNP site. Optimal primer length was selected based on temperature and GC content using Oligo Analyzer 1.0 software (http://oligo-nalyzer.software.informer.com/1.0/).
The nucleotide sequences of these primers were:
(FOV_BT_SNP_R3) 5’-GTGTAGTGACCCTTGGCC CA-3’ and
(FOV_BT_AS_R3) 5’-GTGTAGTGACCCTTGGCCC AA-3’.
PCR using race-specific primers and SNP detection
PCR-amplicons of BT3 and BT5 primer pairs were purified with SAP and ExoI (Affymetrix Inc., Santa Clara, CA). SNP was detected using primer (FOV_BT_SNP_R3) and ABI Prism SNaPshot Multiplex Kit (Applied Biosystems, Foster City, CA) according to manufacturer’s protocol on Genetic Analyzer 3130xl (Applied Biosystems, Foster City, CA). Allele specific PCR was performed using primers FOV_BT_AS_R3 under the following protocol: 95°C for 5 min, and remaining 25 cycles consisted of 94°C for 30 s, 59°C for 15 s, and 72°C for 30 s, with a final extension at 72 °C for 7 min.
Results and discussion
The universal fungal (BT3/BT5) primers generated ~600 bp long PCR products from all tested pathogen isolates. Sequence analysis revealed G/A polymorphism of the amplified fragment of BT gene. Further analyses of sequences showed that all FOV race 3 isolates had nucleotide ‘A’ at position 116 bp (), whereas nucleotide ‘G’ was present in other FOV races (1, 2, 4, 6, 7 and 8). The allele specific primer had a target nucleotide ‘A’ on 3’-end to assure allele-specific annealing of primer. The allele specific forward primer FOV_BT_AS_R3 and universal reverse primer BT-5 were able to clearly discriminate the race 3 from other races of FOV (). The BT gene along with other single-copy genes is used for identification and taxonomic studies of fungi (Baldauf & Palmer Citation1993; Edlind et al. Citation1996; Baldauf & Doolittle Citation1997; O’Donnell et al. Citation1998; Geiser et al. Citation2001; Mach et al. Citation2004; Reischer et al. Citation2004; Yli-Mattila et al. Citation2004). Previously, we aimed to characterize two single-copy genes (e.g. EF, BT), and rDNA of several Uzbek ecotypes of virulent isolates of F. oxysporum f.sp. vasinfectum (Egamberdiev et al. Citation2013) that helped us to assign the race identity of unknown Uzbekistan FOV isolates. Sequence comparisons of known race DNAs and DNAs from unknown isolates detected several FOV race-specific SNPs that were subject of this study.
Fig. 1 SNP discovery after alignment of amplified regions of β-tubulin gene of FOV isolates representing different races. Unique polymorphism intrinsic to race 3 is labeled in bold.
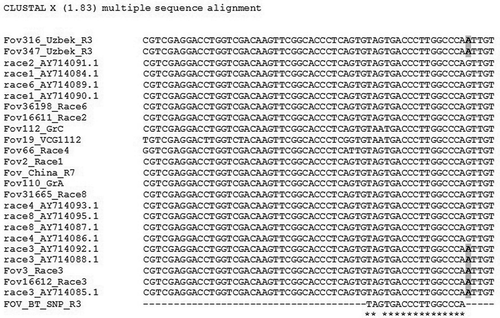
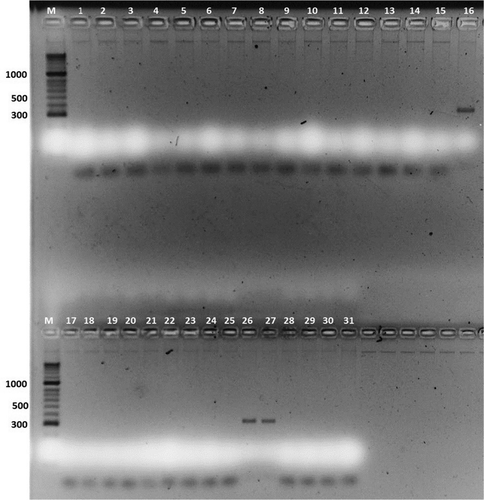
Fig. 2 (Colour online) Allele specific PCR (ASPCR) performed with the primer (FOV_BT_AS_R3) specific for Fusarium oxysporum f. sp. vasinfectum race 3 where: M-HyperLadder II (Bioline USA inc.); 1- 560; 2- 552; 3- 533; 4- 546; 5- 547; 6- FOV 2; 7- FOV 16611; 8- FOV 66; 9- FOV 36198; 10- FOV 7 China; 11- FOV 31665; 12- FOV 19; 13- FOV 110; 14- FOV 124; 15- FOV 112; 16- FOV 3; 17- 532; 18- 403 b; 19- 28; 20- 139; 21- 319; 22- 520; 23- 489; 24- 496; 25- 526; 26- 316; 27- 347; 28- 37529. 444; 30- 328; 31- 532 (see for isolate details).
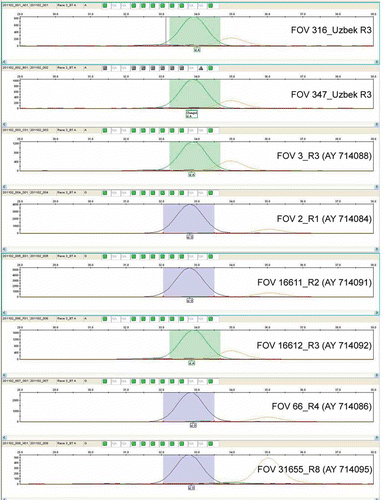
Table 1. Strains of Fusariuum oxysporium f. sp. vasinfectum of known races, Uzbekistan isolates and five species of Fusarium genus used in the AS and ASPCR analyses.
Specificity of ASPCR amplification for FOV using primers BT5 and newly developed FOV_BT_AS_R3 primer pairs were tested on 31 isolates which contain seven races of FOV (8 Uzbek isolates, representing races 1, 3, 4, and 7 as well as known FOV races: 1, 2, 3, 4, 6, 7 and 8, used as control samples), and 5 species of Fusarium genus (). The specificity of primers was tested by adjusting the annealing temperature and DNA concentration. No cross-reaction was detected with genomic DNA from any isolates of other FOV races and Fusarium species. The optimal annealing temperature for ASPCR was 59°C. DNA dilution analysis showed that the optimum concentration of fungal DNA is 5 ng/μL for ASPCR.
Previous reports stated that 50 fg to 100 ng of DNA extracted from F. oxysporum f. sp. vasinfectum mycelium was found to be suitable for PCR (Moricca et al. Citation1998). Detection of FOV from genomic DNAs of diseased parts of plants (root, leaves, seeds) does clearly show FOV race-3 specific amplification products (), where the genomic DNAs were extracted with CTAB method (Dellaporta et al. Citation1983) from infected versus non infected seedlings of ‘Namangan – 77’ cultivar. Thus, PCR with primer set of BT-5 (5’GCT CTAGACTGCTTTCTGGCAGACC -3’ (Tooley et al. Citation2001) and specific FOV_BT_AS_R3 (5’- GTGTAGTGACCCTTGGCCCAA-3’) developed for β-tubulin gene in our study specifically amplified a fragment of 321 bp from all FOV race 3 as well as diseased plant tissues infected with FOV race 3. This demonstrated usefulness of AS primer for rapid and efficient detection of FOV race 3.
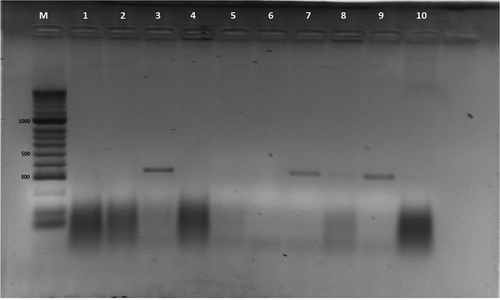
Fig. 3 Detection of FOV from diseased parts of plant (root, stem, leaves, seeds): M-HyperLadder II (Bioline Inc., USA); 1 – non-infected plant root tissue; 2 – non-infected plant leaf tissue; 3 – FOV race 3, a monosporic isolate; 4 – infected plant with FOV race 1; 5 – infected plant with FOV race 4 FOV; 6 – infected plant with root rot (F. solani) isolate; 7 – plant root tissues infected with FOV race 3 (isolate 316); 8–plant seed tissue infected with FOV race 3 (isolate 316); 9–plant root tissue infected with FOV race 3 (isolate 347); and 10 – non-infected seed tissue. Note: sample no. 8 revealed very week, but noticeable amplification product. Additional information for the rest of the samples (11 – 31) can be found in .
Further, on the basis of revealed race-specific polymorphism, a single-base extension SNP genotyping method (ABI Prism SNaPshot Multiplex Kit, Applied Biosytems) was adapted to identify FOV race 3. The developed single-base extension SNP-typing procedure showed a high level of specificity for identification of FOV race 3. We tested the SNP primer (FOV_BT_SNP_R3) against a control set of FOV races and local ecotypes of race 3 including fungal isolates FOV 316 and FOV 347 (). The SNP primer also was able to identify FOV race 3 controls (AY714088 and AY714092) belonging to Californian and Egyptian isolates ().
Fig. 4 Results after capillary electrophoresis using primer FOV_BT_SNP_R3 and ABI Prism SNaPshot Multiplex Kit (Applied Biosystems, Foster City, CA). Peaks marked additionally by green color, showing a presence of SNP (nucleotide A) and detect race 3 specifically.
The newly developed SNP primer coupled with single-base extension (SNaPshot) technique proved to be a robust tool to identify FOV race 3 from different geographic origins. The main advantages of this approach are its accuracy and convenience of data processing. Additionally, the ability to use differently labeled fluorescent ddNTPs allows identification of more than one target nucleotide at a time, giving certain advantages for high throughput experiments. However, the major disadvantages of this method are the high price and the need for expensive equipment to perform the assay. The increasing need for large-scale genotyping applications of SNPs in FOV race identification requires the development of low-cost technologies accessible to minimally equipped laboratories. As an alternative to the SNaPshot technique, the allele specific PCR approach described herein can be used.
In taxonomic analysis, the β-tubulin gene has the following advantages over other single copy genes: 1) insertions and deletions are much less common, making sequence alignments less ambiguous; and 2) the gene has been studied in several ascomycetous fungi (Thon & Royse Citation1999). The newly developed BT allele-specific PCR method for detection of FOV race 3 allows efficient discrimination of SNPs in a single reaction with standard PCR conditions. This approach allows rapid screening of plant pathogen collections for isolates belonging to FOV race 3 since this pathogen is typical to Uzbekistan soils. This is a robust, timesaving, and inexpensive method which requires only a minimally equipped laboratory for fast identification of FOV pathogen, thereby helping to predict plant disease outbreaks.
Acknowledgements
The authors thank the Office of International Research Programs, US Department of Agriculture (USDA) for funding this study under research grant UZB2-31016-TA-09 and U.S. Civilian Research & Development Foundation (CRDF) for project coordination. We thank Dr. Brain E. Scheffler, USDA-ARS, Genomics and Bioinformatics Research Unit for sequencing Fov isolates. We are indebted to Dr. R. Davis, Department of Plant Pathology, Univ. of California, Davis, CA for providing reference FOV race DNAs. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. The U. S. Department of Agriculture is equal opportunity provider and employer.
References
- Abd-Elsalam KA, Aly IN, Abdel-Satar MA, Khalil MS, Verreet JA. 2003. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr J Biotechnol. 2:82–85.
- Abd-Elsalam KA, Asran-Amal A, Schnieder F, Migheli Q, Verreet JA. 2006. Molecular detection of Fusarium oxysporum f.sp. vasinfectum in cotton roots by PCR and real-time PCR assay. J Plant Dis Protect. 113:14–19.
- Abdullaev A, Salahutdinov I, Kuryazov Z, Sh E, Rizaeva S, Ulloa M, Abdurakhmonov IY. 2011. Study on Fusarium wilt disease (F. oxysporum vasinfectum) in Upland cotton (G. hirsutum). In: Fifth World Cotton Research Conference; 2011 Nov 7–11; Mumbai, India.
- Assigbetse KB, Fernandez D, Dubois MP, Geiger JP. 1994. Differentiation of Fusarium oxysporum f.sp. vasinfectum races on cotton by random amplified polymorphic DNA (RAPD) analysis. Phytopathology. 84:622–626. doi:10.1094/Phyto-84-622
- Attitalla IH, Fatehi J, Levenfors J, Brishammar S. 2004. A rapid molecular method for differentiating two special forms (lycopersici and radicis-lycopersici) of Fusarium oxysporum. Mycol Res. 108:787–794. doi:10.1017/S0953756204000322
- Baayen RP, O’Donnell K, Bonants PJM, Cigelnik E, Kroon LPNM, Roebroeck EJA, Waalwijk C. 2000. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic Formae Speciales causing wilt and rot disease. Phytopathology. 90:891–900. doi:10.1094/PHYTO.2000.90.8.891
- Baayen RP, O’Donnell K, Breeuwsma S, Geiser DM, Waalwijk C. 2001. Molecular relationships of fungi within the Fusarium redolens - F. hostae clade. Phytopathology. 91:1037–1044. doi:10.1094/PHYTO.2001.91.11.1037
- Baldauf SL, Doolittle WF. 1997. Origin and evolution of the slime molds (Mycetozoa). Proc Natl Acad Sci. 94:12007–12012. doi:10.1073/pnas.94.22.12007
- Baldauf SL, Palmer JD. 1993. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci. 90:11558–11562. doi:10.1073/pnas.90.24.11558
- Bayraktar H, Dolar FS, Maden S. 2008. Use of RAPD and ISSR markers in detection of genetic variation and population structure among Fusarium oxysporum f.sp. ciceris isolates on chickpea in Turkey. J Phytopathol. 156:146–154. doi:10.1111/j.1439-0434.2007.01319.x
- Beckman CH, Roberts EM. 1995. On the nature and genetic basis for resistance and tolerance to fungal wilt diseases of plants. Adv Bot Res. 21:35–77. doi:10.1016/S0065-2296(08)60008-7
- Bogale M, Wingfield BD, Wingfield MJ, Steenkamp ET. 2006. Characterization of Fusarium oxysporum isolates from Ethiopia using AFLP, SSR and DNA sequence analyses. Fungal Div. 23:51–66.
- Burgess LW. 1981. General ecology of the Fusaria. In: Nelson PE, Toussoun TA, Cook RJ, editors. Fusarium: diseases, biology and taxonomy. University Park, Pennsylvania: Pennsylvania State University Press; pp. 225–235.
- Dellaporta SL, Wood J, Hicks JP. 1983. A plant DNA mini-preparation: Version II. Plant Mol Biol Rep. 1:19–21. doi:10.1007/BF02712670
- Edlund TD, Li J, Visvesvara GS, Vodkin MH, Mclaughlin GL, Katiyar SL. 1996. Phylogenetic analysis of β-tubulin sequences from amitochondrial protozoa. Mol Phylogenet Evol. 5:359–367. doi:10.1006/mpev.1996.0031
- Egamberdiev SS, Ulloa M, SAHA S, Salakhutdinov IB, Abdullaev AA, Glukhova LA,Adylova AT, Scheffler BE, Jenkins JN, Abdurakhmonov IY. 2013. Molecular characterization of Uzbekistan isolates of Fusarium oxysporum f. sp. vasinfectum. J. Plant Sci Mol Breed. [Internet]. [cited 2013 Aug 19]; 2, 3. Available from: http://www.hoajonline.com/Journal-of-Plant-Science-and-Molecular.html. doi:10.7243/2050-2389-2-3
- Fernandez D, Assigbetse KB, Dubois MP, Geiger JP. 1994. Molecular characterization of races and vegetative compatibility groups in Fusarium oxysporum f.sp. vasinfectum. Appl Environ Microbiol. 60:4039–4046.
- Geiser DM, Juba JH, Wang B, Jeffers SN. 2001. Fusarium hostae sp. nov. A relative of F. redolens with a Gibberella teleomorph. Mycologia. 93:670–678. doi:10.2307/3761821
- Kim Y, Hutmacher RB, Davis RM. 2005. Characterization of California isolates of Fusarium oxysporum f sp. vasinfectum. Plant Dis. 89:366–372. doi:10.1094/PD-89-0366
- Lievens B, Rep M, Thomma BPHJ. 2008. Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manag Sci. 64:781–788. doi:10.1002/ps.1564
- Mach RL, Kullnig-Gradinger CM, Farnleitner AH, Reischer G, Adler A, Kubicek CP. 2004. Specific detection of Fusarium langsethiae and related species by DGGE and ARMS-PCR of a β-tubulin (tub1) gene fragment. Int J Food Microbiol. 95:333–339. doi:10.1016/j.ijfoodmicro.2003.12.011
- Marupov A, Boijigitiv F, Irgasheva N. 2012. [Fusarium wilt of cotton]. AgroIlm. 2:39–40. Russian.
- Marupov A, Ishankulova M, Rahmatov A, Kim R. 2010. [About wilt diseases of commercialized cotton varieties]. AgroIlm. 4:11–13. Russian.
- Moricca S, Ragazzi A, Kasuga T, Mitchelson KR. 1998. Detection of Fusarium oxysporum f. sp. vasinfectum in cotton tissue by polymerase chain reaction. Plant Pathol. 47:486–494. doi:10.1046/j.1365-3059.1998.00262.x
- Nazar RN, Hu X, Schmidt J, Culham D, Robb J. 1991. Potential use of PCR-amplified ribosomal intergenic sequences in the detection and differentiation of Verticillium wilt pathogens. Physiol Mol Plant Pathol. 39:1–11. doi:10.1016/0885-5765(91)90027-F
- O’Donnell K, Cigelnik E, Nirenberg HI. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 90:465–493. doi:10.2307/3761407
- O’Donnell K, Kistler HC, Tacke BK, Casper HH. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci. 97:7905–7910. doi:10.1073/pnas.130193297
- Reischer GH, Lemmens M, Farnleitner A, Adler A, Mach RL. 2004. Quantification of Fusarium graminearum in infected wheat by species specific real-time PCR applying a TaqMan probe. J Microbiol Meth. 59:141–146. doi:10.1016/j.mimet.2004.06.003
- Skovgaard K, Nirenberg HI, O’Donnell K, Rosendahl S. 2001. Evolution of Fusarium oxysporum f.sp. vasinfectum races inferred from multigene genealogies. Phytopathology. 91:1231–1237. doi:10.1094/PHYTO.2001.91.12.1231
- Thon MR, Royse DJ. 1999. Partial beta-tubulin gene sequences for evolutionary studies in the Basidiomycotina. Mycologia. 91:468–474. doi:10.2307/3761347
- Tooley PW, Goley ED, Carras MM, Frederick RD, Weber EL, Kuldau GA. 2001. Characterization of Claviceps species pathogenic on sorghum by sequence analysis of the β-tubulin gene intron 3 region and EF-1α gene intron 4. Mycologia. 93:541–551. doi:10.2307/3761739
- Ulloa M, Hutmacher RB, Roberts PA, Wright SD, Nichols RL, Davis RM. 2013. Inheritance and QTL mapping of Fusarium wilt race 4 resistance in cotton. Theor Appl Genet. 126:1405–1418. doi:10.1007/s00122-013-2061-5
- Yli-Mattila T, Mach RL, Alekhina IA, Bulat SA, Koskinen S, Kullnig-Gradinger CM, Kubicek CP, Klemsdal SS. 2004. Phylogenetic relationship of Fusarium langsethiae to Fusarium poae and Fusarium sporotrichioides as inferred by IGS, ITS, β-tubulin sequences and UP-PCR hybridization analysis. Int J Food Microbiol. 95:267–285. doi:10.1016/j.ijfoodmicro.2003.12.006
- Zambounis AG, Paplomatas E, Tsaftaris AS. 2007. Intergenic spacer–RFLP analysis and direct quantification of Australian Fusarium oxysporum f.sp. vasinfectum isolates from soil and infected cotton tissues. Plant Dis. 91:1564–1573. doi:10.1094/PDIS-91-12-1564
