Abstract
Abstract: Peach yellows disease was identified in a visual survey conducted in Shaanxi, China in 2010 and 2011. Using transmission electron microscopy and a nested PCR assay, the disease was confirmed to be due to phytoplasma infection. Based on sequence data, phylogenetic analysis and restriction fragment length polymorphism (RFLP) analysis, the peach yellows phytoplasma was classified into the subgroup 16SrV-B. Bud graft with fresh buds from diseased peach trees as scions onto healthy young peach trees as rootstocks showed that the phytoplasma was a causal agent of peach yellows disease and was graft-transmissible. This is the first reported occurrence of 16SrV-B phytoplasma in peach trees in northwest China.
Résumé
La jaunisse du pêcher a été identifiée au cours d’une étude visuelle menée en 2010 et 2011 dans la province du Shaanxi, en Chine. La maladie, une phytoplasmose, a été confirmée par microscopie électronique à transmission et PCR par amorces incluses. En raison des données de la séquence, de l’analyse phylogénétique et du polymorphisme de longueur des fragments de restriction (RFLP), le phytoplasme de la jaunisse du pêcher a été classé dans le sous-groupe 16SrV-B. Des greffons provenant de pêchers infectés, greffés sur de jeunes pêchers sains, ont servi à démontrer que le phytoplasme était l’agent causal de la jaunisse du pêcher et qu’il était transmissible par greffe. Il s’agit du premier cas de phytoplasme 16SrV-B rapporté sur les pêchers dans le nord-ouest de la Chine.
Introduction
The peach tree (Prunus persica L. Batsch), which is native to China, is grown on more than 7670 km2, with a production of about 11.5 million metric tons annually in China (FAO Citation2011). A yellows disease in peach trees caused by phytoplasmas has been found in Jordan, India, south China and Bolivia, and is associated with the phytoplasmas belonging to groups 16SrI (Anfoka & Fattash Citation2004), 16SrV (Lee et al. Citation2004; Huang et al. Citation2009) and 16SrXII (Jones et al. Citation2005).
Shaanxi, in northwest China, is one of the most important areas for crop and fruit production, mainly of wheat, kiwi, apple, jujube and peach. Phytoplasma diseases seriously reduce yields, and Shaanxi has been affected by several diseases. Wheat blue dwarf (WBD) disease, for example, caused by the phytoplasmas of subgroup 16SrI-C spread to an area of 90 000 km2 and caused losses of c. 50 000 metric tons in 1960 (Wu et al. Citation2010). Jujube witches’ broom disease, caused by the phytoplasmas of the subgroup 16SrV-B, resulted in a large number of jujube trees being cut down every year (Tian Citation1998). Several other important phytoplasma diseases have been diagnosed in Shaanxi, such as Puna chicory flat stem (Li, Zhang, Bai et al. Citation2012), plum leaf roll (Min et al. Citation2011) and apricot leaf roll (Yue et al. Citation2009).
The objective of this study was to confirm the occurrence of phytoplasma in peach yellows-affected peach trees using transmission electron microscopy and PCR-restriction fragment length polymorphism (RFLP) analysis. Results presented show that the phytoplasma was a causal agent of the peach yellows disease, which was confirmed with a bud graft test.
Materials and methods
Sample collection
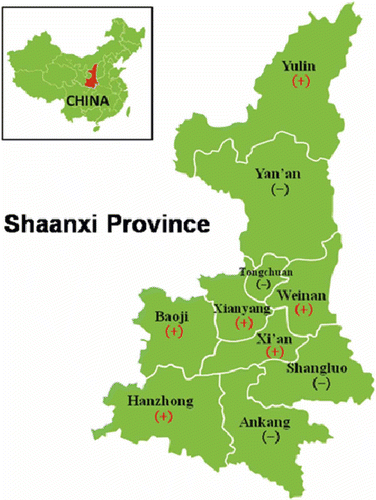
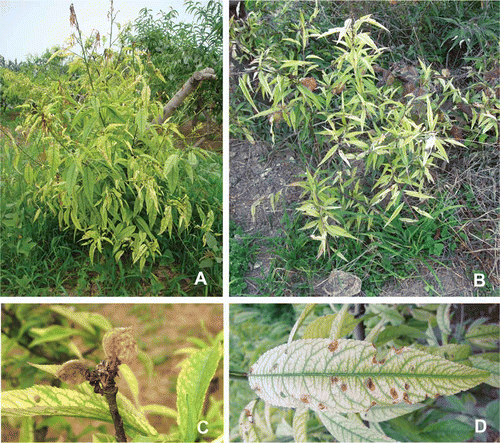
From 2010 to 2011, visual surveys were conducted between June and September in commercial peach orchards (about 70 km2 total area) in a number of cities, distributed from north to south in Shaanxi (). We observed peach yellows disease at six sites. Disease symptoms included yellowing, upward curling, necrotic and tattered leaves, small and wilted fruit, growth decline and dieback (). Sixty symptomatic young branches were sampled from diseased trees at six sites (), and were immediately stored in an ice cooler box and transported back to the laboratory. Samples were kept at −86 °C for PCR, and the cut fresh tissues were dipped into a fixative solution for transmission electron microscopy (TEM) analysis. Asymptomatic branch samples were also sampled as negative controls at the six sites. Dry plant materials stored at −86 °C served as positive controls, and included jujube witches’ broom (JWB) phytoplasmas (16SrV-B), chinaberry witches’ broom (CWB) phytoplasmas (16SrI-B) and mulberry dwarf (MD) phytoplasmas (16SrI-B).
Transmission electron microscopy
Small rectangular segments (2 × 2 mm2) containing the central midrib of fresh, symptomatic leaves and healthy controls were fixed in a mixture of 3% (v/v) glutaraldehyde and 4% (v/v) paraformaldehyde for 4 h at 4 °C, followed by 1% (w/v) osmium tetroxide for 2 h at room temperature. After dehydration with ethanol in serial dilutions from 10% to 70% (v/v) (Kamińska et al. Citation2001), the samples were embedded in Epon812 before cutting with a diamond knife. The ultrathin sections (70 nm) were supported by 400 mesh copper grids, stained with 5% uranyl acetate for 30 min and lead citrate for 10 min, and observed under a HT7700 TEM (Li, Zhang, Zhao et al. Citation2012).
DNA extraction and polymerase chain reaction (PCR)
Total DNA was isolated from 0.2 g leaf samples using the cetyltrimethyl ammonium bromide (CTAB) method (Angelini et al. Citation2001) and served as a template in nested PCR amplification, which targeted the partial 16S rRNA gene of phytoplasmas. In two runs of the nested PCR, the reaction mixture components (25 µL) were set as 20 ng DNA template for the first run, and dilution of 1 : 30 of first products for the second run, 2.0 µL of 10 µM primer pairs, 2.0 μL of 2.5 mM deoxynucleotide triphosphate, 2.5 μL of 25 mM MgCl2, 2.5 μL of 10 × Taq DNA polymerase buffer and 0.2 μL Taq DNA polymerase (TaKaRa Bio Inc., Dalian, China). The two sequential sets of primers were P1/P7 (Deng & Hiruki Citation1991) for the first reaction and R16F2n/R16R2 (Lee et al. Citation1998) for the second reaction.
The thermal parameters for the first run were pre-denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 2 min, and extension at 72 °C for 1 min. Finally, an extra 10 min extension was performed. For the second run, only the annealing temperature was changed to 52 °C. The first and second PCR products were separated in 1% agarose gel by electrophoresis, after staining with ethidium bromide visualized under ultraviolet light.
Sequence analysis
All R16F2n/R16R2 amplicons were purified with PCR Products Purification Kit (Bio Teke Co., Beijing, China) and ligated into pMD18-T simple vectors (TaKaRa Bio Inc., Dalian, China). The recombinant plasmids, which were finally sequenced by TaKaRa Bio Inc. (Dalian, China), were then transformed into the Escherichia coli strain JM109 for efficient replication. Cloned plasmids were isolated using a Plasmid Miniprep Kit (Sangon Biotech Co., Ltd., Shanghai, China). For each sample, three positive single colonies were randomly selected and cultured to isolate the plasmids. The BLASTn tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search homogeneous sequences matching the query sequences in the NCBI nucleotide database. The identities between the query sequences and its homogeneous sequences were calculated with pairwise comparison using DNAMAN software, version 6.0 (Lynnon Biosoft, QC, Canada), and homogeneous sequences were flanked with primers R16F2n/R2. A phylogenetic analysis was carried out with the query sequences and previously published sequences of phytoplasmas (). The phylogenetic tree was generated with the neighbour-joining method using a 1000-replicate bootstrap search and MEGA5 software (Tamura et al. Citation2011).
Table 1. 16SrRNA gene sequences of phytoplasmas used for the phylogenetic analysis.
RFLP analysis
In accordance with the manufacturer’s instructions (TaKaRa Bio Inc., Dalian, China), the second products of nested PCR (in volume of 25 μL; in concentration range of 50–70 ng μL−1) were digested with seven restriction enzymes (AluI, BfaI, EcoRI, HhaI, HpaII, MseI and RsaI) recognized from published RFLP patterns. This allowed us to differentiate subgroups of 16SrV as well as groups of 16Sr (Lee et al. Citation1998). The digested fragments were then separated in a vertical 8% polyacrylamide gel and visualized under UV after staining with ethidium bromide.
An in silico RFLP analysis was performed for the obtained sequences with the online tool iPhyClassifier (http://www.ba.ars.usda.gov/data/mppl/iPhyClassifier.html). Seventeen enzymes were employed for digestion, containing AluI, BamHI, BfaI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI and HpaII. KpnI, MseI, RsaI, Sau3AI, SspI and TaqI, and virtual gel images were generated.
Graft transmission
A bud graft transmission test was carried out in July 2010 using three healthy one-year-old peach trees with buds from phytoplasma-infected peach trees as scions. The healthy peach trees as rootstocks were grown from seeds in an insect-free greenhouse. They were confirmed as phytoplasma-free through nested PCR, as described above. The scions, which were PCR-positive for phytoplasmas, were collected in Xi’an. For each tree, one scion was grafted using the method described by Abou-Jawdah et al. (Citation2003). The young, grafted peach trees were first kept for three days at a shady location, such as under greenhouse benches, and protected from drying out. Later, they were grown on greenhouse benches at about 24 °C with a relative humidity of around 60%. The greenhouse was kept free from insects using mesh. Phytoplasmas were detected by nested PCR and RFLP, as described above in July 2011, after symptoms of yellows developed.
Results
PCR detection
In the nested PCR, the first and second products of size c. 1.8 and 1.2 kb, respectively, were obtained. The 1.8 kb bands, as seen from a gel image (not shown), originated from some samples collected in Baoji, Weinan and Xi’an. Bands above 1.2 kb were obtained from other samples (). From samples of positive controls, 1.8 and 1.2 kb fragments were obtained. No product was obtained from asymptomatic samples used as negative controls. Based on the results of PCR detection, 35% of the total collected samples were confirmed to have peach yellows disease.
Table 2. Results of nested PCR detection for phytoplasma in peach.
Table 3. Identities between the nine sequences determined in this study and its homogeneous sequences.
TEM observation
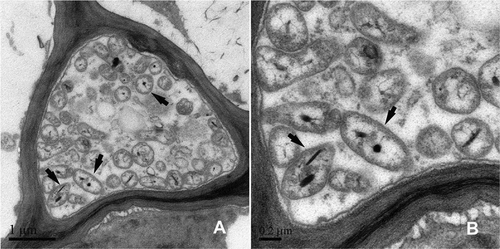
Using TEM, we observed many phytoplasma cells in the phloem of peach trees exhibiting yellows symptoms, but none in healthy peach trees (). The phytoplasmas were characterized as irregular, spherical in shape with a diameter ranging from 400 to 600 nm. The particles in high density near the membrane were presumed to be ribosomes and the fibrillar material in the central region was presumed to contain DNA.
Sequence analysis
We obtained nine different sequences of 16S rRNA genes of phytoplasmas labelled as PYaP–Yangling (YL), Weinan (WN), Hanzhong (HZ), Xi’an (XA)1 and XA2, Xianyang (XY)1 and XY2, Baoji (BJ)1 and BJ2. They were deposited in GenBank under accession numbers KF523369–523377. The identities of the nine sequences, by pair-wise comparison, ranged from 99.6% to 99.8% (). In further analysis, the nine sequences were compared, pair-wise, with representative sequences from the NCBI database, the peach yellows phytoplasmas in India (accession numbers AY197660 and AY332658), the phytoplasmas of 16SrV-B in Shaanxi (accession numbers FJ459914 and DQ919060) and the ‘Ca. Phytoplasma ziziphi’ strain JWB-G1 (AB052876), respectively . Most of the nine sequences were highly homologous with FJ572660 (Apricot leaf roll phytoplasma) and HQ844237 (Golden bellflower yellows phytoplasma). All nine were the least homologous with EF432569 (Bischofia polycarpa witches’-broom phytoplasma). The pair-wise identities between the nine and AY197660 or AY332658 (Indian peach yellows phytoplasma) ranged from 99.52% to 99.68%. For the published sequences of subgroup 16SrV-B in Shaanxi, the identities were 99.6–99.8% to FJ459914 (plum roll leaf phytoplasma), and 98.64–98.8% to DQ919060 (jujube witches’ broom phytoplasma). For AB052876 (JWB-G1), the identity range was from 99.59% to 99.67%.
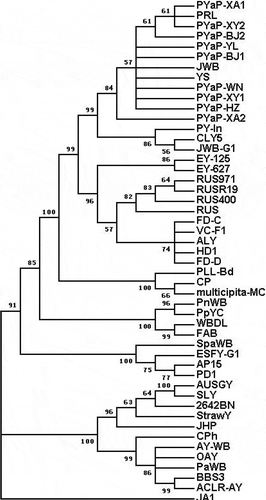
We selected a total of 49 sequences of phytoplasmas, including the nine query sequences and an outgroup sequence, from six phytoplasmal groups of 16SrI, -II, -V, -VI, -X and -XII and used them for phylogenetic analysis after they were flanked with the primers R16F2n/R2. A phylogenetic tree was inferred (), in which the nine query sequences clustered together and showed closely phylogenetic relationships with published sequences of phytoplasmas in the subgroup 16SrV-B. These contained AY197660 from peach yellows phytoplasmas in India, DQ919060 from jujube witches’ broom phytoplasmas, AY197659 from cherry lethal yellows phytoplasmas, FJ572660 from apricot leaf roll phytoplasmas and FJ459914 from plum roll leaf phytoplasmas.
RFLP analysis
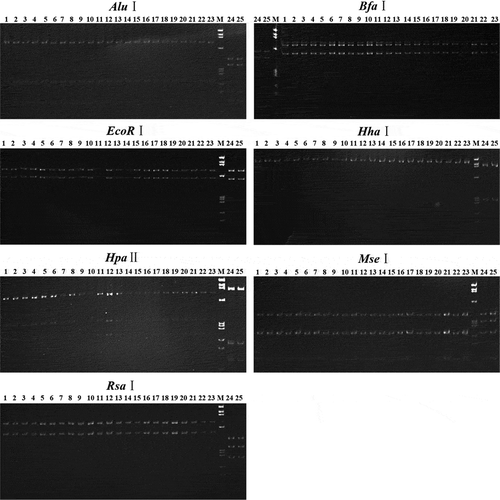
As shown in the RFLP patterns (), the 1.2 kb nested PCR products from the 21 (out of 60) symptomatic samples, noted as numbers 1 to 21, generated the same discrete DNA bands after digesting with the enzymes AluI, BfaI, EcoRI, HhaI, HpaII, MseI and RsaI. Moreover, DNA bands from the 21 samples were found to be identical to those from the JWB phytoplasmas (subgroup 16SrV-B) (labelled with number 23) in size; however, they were distinct from those in CWB and MD (subgroup 16SrI-B) marked with numbers 24 and 25, respectively.
Fig. 5 Separation of nested PCR products after digesting with enzymes. Lanes 1–21, PYaP. Lane 22, graft-transmitted PYaP. Lane 23, JWB. Lanes 24, CWB. Lane 25, MD. Lane M, DNA marker ΦX174 HaeIII digest DNA ladder; band sizes from top to bottom are 1353, 1078, 872, 603, 310, 281, 271, 234, 194 and 118 bp.
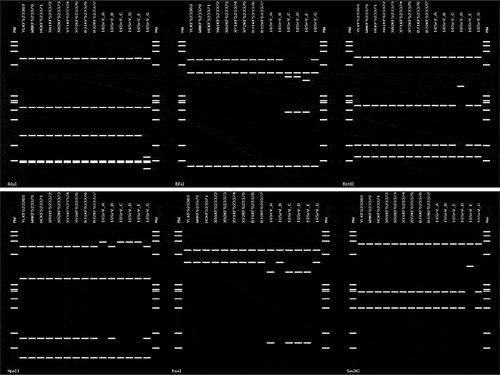
Furthermore, the patterns generated by iPhyClassifier indicated that, after digestion with 17 enzymes, the nine sequences from peach yellows phytoplasmas generated the same patterns, which were also identical to those of subgroup 16SrV-B. However, these patterns differed from those in subgroups 16SrV-A, -C, -D, -E and -G for enzymes AluI, BfaI, BstUI, HpaII, RsaI and Sau3AI (). Therefore, we classified the phytoplasmas associated with peach yellows disease into subgroup 16SrV-B.
Graft transmission
In late May 2011, some young leaves of the three grafted peach trees began to yellow, with all leaves yellowing in July. The symptoms were quite similar to those of the naturally phytoplasma-infected peach trees (). The results of a nested PCR-RFLP analysis confirmed the infection of phytoplasmas of subgroup 16SrV-B in these young trees; the separation of digested DNA bands is shown for number 22.
Discussion
Diagnosis of diseases caused by phytoplasmas is difficult when the detection is based on field symptoms. However, PCR is a sensitive method for detecting phytoplasma pathogens. Based on the main symptom of yellows, we collected 60 samples from diseased peach trees and symptomatically inferred that the disease was associated with phytoplasmas. Nested PCR assays were then conducted to detect phytoplasma presence in the samples. Based on the results, 21 of 60 samples were confirmed as PCR-positive for phytoplasma infection. This implied that there were other casual agents of yellows in peach trees. Previous research reports indicate that iron deficiency can also cause yellowing in plants (Ren et al. Citation2009).
Next, we diagnosed 60 symptomatic samples with nested PCR, and found that 21 samples were positive in the second run, as the products of c. 1.2 kb were generated. However, only eight samples (out of the 21) were positive in the first run, according to the DNA bands of c. 1.8 kb on a 1% agarose gel. Given that the second run of the nested PCR used the diluted first products as templates, we considered that the phytoplasmas occurred in a low titre in most of the infected samples (Zirak et al. Citation2010) or that there were some PCR inhibitors in the total DNA (Lepka et al. Citation1999; Musetti et al. Citation2000; Heinrich et al. Citation2001).
Using bud graft transmission, in which fresh buds of phytoplasma-infected peach trees are used as scions and healthy young peach trees as rootstocks, we confirmed the phytoplasmas (PYaP) as the causal agent of the peach yellows disease, which was graft-transmissible. Since grafting is a main method of breeding high-quality peach trees, graft-transmissible phytoplasma could be a potential threat to peach production.
Finally, we searched the published phytoplasma-associated diseases in peach trees. A significant phytoplasma of group 16SrX, called European stone fruit yellows (ESFY) phytoplasma, has been found to cause severe damage in European peach trees (Seemuller & Schneider Citation2004). Another important phytoplasma of 16SrIII has been found to induce peach X disease in North America (Wang et al. Citation2012; Davis et al. Citation2013). Additionally, phytoplasmas of 16SrI-W and 16SrVII have been reported in Canada (Arocha-Rosete et al. Citation2011), and phytoplasmas of 16SrII, -V, -VI, -IX and -XII infected peach trees have been found in Asia (Verdin et al. Citation2003; Lee et al. Citation2004; Zirak et al. Citation2010). In China, the phytoplasmas of subgroup 16SrI-B and -C have been found to cause peach red leaf disease (Zhang et al. Citation2005, Citation2013), while another peach yellows disease has also been reported (Huang et al. Citation2009) in Sichuan, south China. This peach yellows disease has been associated with the phytoplasmas of subgroup 16SrV-B. However, sequence data were not available for that study. To our knowledge, our study constitutes the first reported occurrence of phytoplasmas of subgroup 16SrV-B causing peach yellows disease in northwest China. It is our hope that this research will contribute to the eventual eradication of this disease, improving peach production in China and throughout the world.
Acknowledgements
We thank Paul Randall and Eunice Randall of Agriculture and Agri-Food Canada, Pacific Agri-Food Research Center, Canada, for revisions and suggestions to the manuscript.
Funding
ZhengNan Li is a recipient of a China Scholarship Council PhD fellowship. This work was supported by the 111 project from the Education Ministry of China [Grant no. B07049], National Natural Science Foundation of China [Grant no. 31371913] and PhD Student Academic Newcomer Award of China Ministry of Education [Grant no. Z107021202].
References
- Abou-Jawdah Y, Dakhil H, El-Mehtar S, Lee IM. 2003. Almond witches’-broom phytoplasma: a potential threat to almond, peach, and nectarine. Can J Plant Pathol. 25:28–32. doi:10.1080/07060660309507046
- Anfoka G, Fattash I. 2004. Detection and identification of aster yellows (16Sr I) phytoplasma in peach trees in Jordan by RFLP analysis of PCR amplified products (16S rDNAs). J Phytopathol. 152:210–214. doi:10.1111/j.1439-0434.2004.00831.x
- Angelini E, Clair D, Borgo M, Bertaccini A, Boudon-Padieu E. 2001. Flavescence doree in France and Italy-Occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis. 40:79–86.
- Arocha-Rosete Y, Zunnoon-Khan S, Krukovets I, Crosby W, Scott J, Bertaccini A, Michelutti R. 2011. Identification and molecular characterization of the phytoplasma associated with peach rosette-like disease at the Canadian Clonal Genebank based on the 16S rRNA gene analysis. Can J Plant Pathol. 33:127–134. doi:10.1080/07060661.2011.558854
- Davis RE, Zhao Y, Dally EL, Lee IM, Jomantiene R, Douglas SM. 2013. Candidatus Phytoplasma pruni’, a novel taxon associated with X-disease of stone fruits, Prunus spp.: multilocus characterization based on 16S rRNA, secY, and ribosomal protein genes. Int J Syst Evol Microbiol. 63:766–776. doi:10.1099/ijs.0.041202-0
- Deng S, Hiruki C. 1991. Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. J Microbiol Meth. 14:53–61. doi:10.1016/0167-7012(91)90007-D
- [FAO] Food and Agriculture Organization of the United Nation. FAOSTAT Database on Production [Internet]. 2011. [cited 2012 May 18]. Available from: http://faostat.fao.org/
- Heinrich M, Botti S, Caprara L, Arthofer W, Strommer S, Hanzer V, Katinger H, Bertaccini A, Machado MLDC. 2001. Improved detection methods for fruit tree phytoplasmas. Plant Mol Biol Rep. 19:169–179. doi:10.1007/BF02772160
- Huang Y, Wang J, Zhao Y, He M. 2009. Identification and sequence analysis of 16S ribosomal DNA of the phytoplasma sp. associated with peach yellow disease. Acta Hortic Sin. 39:341-346. (in Chinese).
- Jones P, Arocha Y, Antesana O, Montilliano E, Franco P. 2005. First report of an isolate of ‘Candidatus Phytoplasma australiense’ associated with a yellow leaf roll disease of peach (Prunus persicae) in Bolivia. Plant Pathol. 54:558. doi:10.1111/j.1365-3059.2005.01234.x
- Jung HY, Sawayanagi T, Shigeyuki K, Nishigawa H, Wei W, Oshima K, Miyata S, Ugaki M, Hibi T, Namba S. 2003. ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’-broom disease. Int J Syst Evol Microbiol. 53:1037–1041. doi:10.1099/ijs.0.02393-0
- Kaminska M, Dziekanowska D, Rudzinska-Langwald A. 2001. Detection of phytoplasma infection in rose, with degeneration symptoms. J Phytopathol. 149:3–10. doi:10.1046/j.1439-0434.2001.00554.x
- Lee IM, Gundersen-Rindal DE, Davis RE, Bartoszyk IM. 1998. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int J Syst Bacteriol. 48:1153–1169. doi:10.1099/00207713-48-4-1153
- Lee IM, Martini M, Marcone C, Zhu SF. 2004. Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ’Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int J Syst Evol Microbiol. 54:337–347. doi:10.1099/ijs.0.02697-0
- Lepka P, Stitt M, Moll E, Seemüller E. 1999. Effect of phytoplasmal infection on concentration and translocation of carbohydrates and amino acids in periwinkle and tobacco. Physiol Mol Plant Pathol. 55:59–68. doi:10.1006/pmpp.1999.0202
- Li ZN, Zhang L, Bai YB, Liu P, Wu YF. 2012. Detection and identification of the elm yellows group phytoplasma associated with Puna chicory flat stem in China. Can J Plant Pathol. 34:34–41. doi:10.1080/07060661.2012.659679
- Li ZN, Zhang L, Zhao L, Wu YF. 2012. A new phytoplasma associated with witches’-broom on Japanese maple in China. For Pathol. 42:371–376. doi:10.1111/j.1439-0329.2012.00769.x
- Min H, Zhang C, Li Z, Zhang J, Zhao Z, Song J, Wu Y. 2011. Identification of elm yellows phytoplasma in plum trees in China. J Phytopathol. 59:57–59.
- Musetti R, Favali M, Pressacco L. 2000. Histopathology and polyphenol content in plants infected by phytoplasma. Cytobios. 102:133–147.
- Ren Y, Jiang Y, Zhai B. 2009. Diagnosis and remedy of the nectarine chlorosis in greenhouse. J Northwest A & F Univ. 37:99-104. (in Chinese).
- Seemuller E, Schneider B. 2004. Candidatus Phytoplasma mali’, ‘Candidatus Phytoplasma pyri’ and ‘Candidatus phytoplasma prunorum’, the causal agents of apple proliferation, pear decline and European stone fruit yellows, respectively. Int J Syst Evol Microbiol. 54:1217–1226. doi:10.1099/ijs.0.02823-0
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi:10.1093/molbev/msr121
- Tian G. 1998. Prevention and treatment strategies of jujube witch’s broom. For Sci Tech. 2:14–16. (in Chinese).
- Verdin E, Salar P, Danet JL, Choueiri E, Jreijiri F, Zammar SE, Gélie B, Bové JM. 2003. Candidatus Phytoplasma phoenicium’ sp nov., a novel phytoplasma associated with an emerging lethal disease of almond trees in Lebanon and Iran. Int J Syst Evol Microbiol. 53:833–838. doi:10.1099/ijs.0.02453-0
- Wang L, Hong N, Martini M, Wang Y, Michelutti R, Wang G. 2012. Molecular characterization of two phytoplasma strains associated with X disease in peach in Canada. Phytopath Mollic. 2:9–14. doi:10.5958/j.2249-4669.2.1.006
- Wu Y, Hao X, Li Z, Gu P, An F, Xiang J, Wang H, Luo Z, Liu J, Xiang Y. 2010. Identification of the phytoplasma associated with wheat blue dwarf disease in China. Plant Dis. 94:977–985. doi:10.1094/PDIS-94-8-0977
- Yue HN, Sun RH, Wei T, Wu YF. 2009. First report of a 16SrⅤ-B group phytoplasma associated with a leafroll-type disease of apricots in northern China. J Plant Pathol. 91:500.
- Zhang H, Cai H, Chen H, Ge X. 2005. Detection and identification of phytoplasma associated with peach red leaf. Plant Quar. 19:7–11. (in Chinese).
- Zhang L, Li ZN, Zhang HW, Tao Y, Wu YF. 2013. Detection and identification of aster yellows group phytoplasma (16SrI-C) associated with peach red leaf disease. J Phytopathol. 161:359–362. doi:10.1111/jph.12064
- Zirak L, Bahar M, Ahoonmanesh A. 2010. Molecular characterization of phytoplasmas associated with peach diseases in Iran. J Phytopathol. 158:105–110. doi:10.1111/j.1439-0434.2009.01585.x