Abstract
A leaf spot disease was observed on cabbage (Brassica oleracea L. var. capitata) affecting 80% of plants growing in greenhouses and fields in Serdang, Selangor, Malaysia. Symptomatic leaf samples were collected from infected plants and isolations made on agar medium. Single-spore isolates from resulting colonies were identified based on cultural and morphological characteristics as Pithomyces chartarum. Morphological identification was confirmed by sequence analysis of the internal transcribed spacer (ITS) regions 1 and 2, including 5.8S rDNA (ITS1-5.8S-ITS2). Pathogenicity tests indicated that P. chartarum causes leaf spot on cabbage. This is the first report of leaf spot caused by P. chartarum on cabbage in Malaysia.
Résumé
Une tache des feuilles a été observée sur le chou (Brassica oleracea L. var. capitata), touchant 80 % des plants poussant en serre et en champ à Serdang, dans l’État du Selangor, en Malaisie. Des échantillons de feuilles symptomatiques ont été collectés sur des plants infectés, après quoi ils ont été isolés sur un milieu de culture à base d’agar. Des isolats de spores uniques, dérivés des colonies qui en ont résulté, ont été identifiés comme Pithomyces chartarum sur la base des caractéristiques culturales et morphologiques. L’identification morphologique a été confirmée par l’analyse de la séquence de l’espaceur transcrit interne (ITS) des régions 1 et 2, y compris l’ITS 5.8S de l’ADNr (ITS1-5.8S-ITS2). Des tests de pathogénicité ont indiqué que P. chartarum cause la tache des feuilles chez le chou. Il s’agit de la première mention de la tache des feuilles causée par P. chartarum sur le chou en Malaisie.
Introduction
Cabbage (Brassica oleracea L. var. capitata) is an economically important crop grown worldwide in approximately 150 countries (FAO Citation2013). This plant is one of the most important vegetable crops cultivated in Malaysia (Nazerian et al. Citation2011). In a February 2012 survey, extensive leaf spot was observed on leaves of cabbage plants that had been cultivated in four greenhouses and two fields in Serdang, Selangor, Malaysia. The disease incidence on cabbage plants was more than 80%. Symptoms of the disease initially appeared as small circular to irregular necrotic spots of grey to blackish colour with bright yellow halos, which were scattered over the leaf surface. As the lesions matured, spots coalesced, forming large necrotic patches. Eventually, severely infected plants defoliated prematurely. To date, no similar disease on cabbage has been reported. The purpose of the present study was to identify the causal agent of the leaf spot on the cabbage plants in Serdang, Selangor, Malaysia.
Materials and methods
Isolation and characterization of the pathogen
In February 2012, diseased leaves with typical symptoms of leaf spot were collected from six infected plants from four greenhouses and two fields of cabbage in Serdang, Selangor, Malaysia. Diseased leaf tissues were cut into 3–5 mm pieces and surface-sterilized in 1% NaOCl for 2 min, rinsed in sterile water twice and dried on sterilized filter paper. A total of 30 pieces were placed on potato dextrose agar (PDA) plates and incubated at 25 °C with a 12 h photoperiod for 10 days. The frequency of recovery of isolates from plated tissues was 100%. A total of six morphologically similar isolates were purified by single spore culturing and subcultured on PDA at 25 °C with a 12 h photoperiod for 10 days, in order to characterize fungal growth and morphology. Shape, length and width of 100 conidia and conidiophores were examined by microscopy.
Pathogenicity test
To determine pathogenicity, four potted 45-day-old healthy plants of cabbage (Brassica oleracea L. var. capitata) ‘Gianty’ were spray inoculated with a conidial suspension containing 1 × 105 conidia mL−1 from 10-day-old sporulating cultures of isolate LCN-S01 (Verma et al. Citation2008). Four plants were also inoculated with sterile distilled water as a control. All plants were initially covered with polythene bags in a greenhouse at 29 °C (day)/23 °C (night) for 48 h. Then, the polythene bags were removed and the plants were kept in the same greenhouse under natural daylight conditions. After 2 weeks, pathogenicity of the isolate was assessed by presence or absence of symptoms of leaf spot. The pathogen was re-isolated and compared with the original isolate. The experiment was repeated three times for confirmation of the results during July–August 2012.
Molecular identification
To confirm the results from morphological studies, the six isolates collected from Serdang, Selangor were grown on PDA and genomic DNA was extracted using the 3% SDS method as described by Gonzlez-Mendoza et al. (Citation2010). The internal transcribed spacer (ITS) regions 1 and 2 and the 5.8S ribosomal DNA (rDNA) region of the isolates were amplified using ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al. Citation1990). The amplified products were resolved by electrophoresis in a 1% agarose gel. The PCR products were purified using Gene JET TM commercial PCR Purification Kit (Fermentas, Axon Scientific, Malaysia) and sequenced using a commercial sequencing service provider (First Base Laboratories Sdn. Bhd., Selangor, Malaysia). Sequences of the ITS-5.8S rDNA were searched in the database of NCBI (http://www.ncbi.nlm.nih.gov). The six sequences from this study and other species of Leptosphaerulina (teleomorphic state of Pithomyces) retrieved from GenBank were aligned using Clustal W (Thompson et al. Citation1994), and manually adjusted as required. Phylogenetic analysis was constructed utilizing the Maximum Likelihood method, in the Jukes–Cantor model (MEGA version 5), with a bootstrap of 1000 replicates (Tamura et al. Citation2011). Trichoderma reesei E.G. Simmons was used as the out-group taxon.
Results and discussion
Isolation and characterization of the pathogen
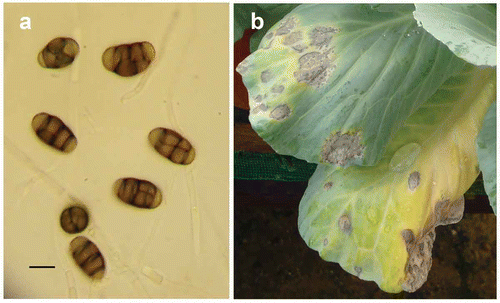
Colonies of the six isolates were dark grey, fast growing, and produced dark brown multicellular conidia on small peg-like branches of the vegetative hyphae. Conidia were broadly elliptical, pyriform and verrucose, measuring 18.0–20.4 × 9.6–12.0 µm (a). Conidiophores were formed laterally on hyphae, measuring 2.4–9.6 × 1.6–3.0 µm. The cultural and morphological characteristics of the isolates were similar to the original description of Pithomyces chartarum (Berk. & M.A. Curtis) M.B. Ellis (Ellis Citation1971).
Pathogenicity test
Symptoms of leaf spot were observed in all of the inoculated cabbage plants, but not in the control plants. The symptoms on the inoculated plants were similar to those observed in the greenhouses and fields. Symptoms consisted of small circular to irregular, grey to blackish spots with bright yellow halos scattered on the leaves. As the lesions matured, spots coalesced together, forming large necrotic patches (b). The pathogen was re-isolated and was found to be identical to the original isolate. The results revealed that P. chartarum was the causal agent of the disease.
Molecular identification
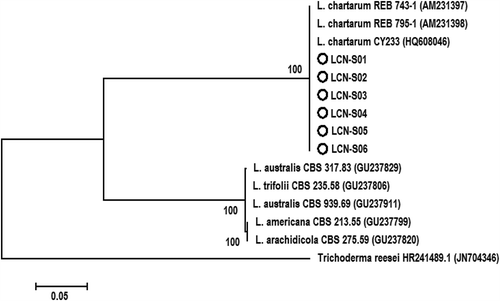
PCR amplification of the ITS-5.8S rDNA region for the six isolates generated a predicted size of 583 bp (). The searches revealed that the six sequences shared 100% identity with each other and Leptosphaerulina chartarum Cec. Roux (teleomorphic state of P. chartarum) (Accession No. HQ608046). The sequence of one representative isolate (LCN-S01) was deposited in GenBank (Accession No. KF951573). In the phylogenetic analysis, the isolates were clustered in a distinct clade with those of L. chartarum isolates retrieved from GenBank with 100% bootstrap value (). These results confirmed the identity of the fungus as P. chartarum.
Fig. 2. Agarose gel showing amplification of the ITS-5.8S rDNA fragments from isolates of Pithomyces chartarum. Lanes 1–6: isolates of P. chartarum. M: 1kb DNA Ladder (Fermentas, Axon Scientific, Malaysia).
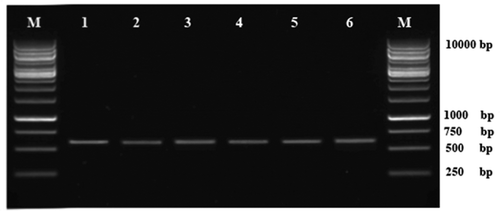
Fig. 3. Phylogenetic tree constructed with the ITS-5.8S rDNA sequence of the six isolates from this study (LCN-S01-LCN-S06), and other species of Leptosphaerulina retrieved from GenBank. Trichoderma reesei was used as the out-group taxon. The bar indicates nucleotide substitutions per site. Numbers of bootstrap support values ≥ 50% based on 1000 replicates.
Pithomyces chartarum is a cosmopolitan, usually saprophytic fungus, but it can cause diseases on a wide range of plants (Ellis Citation1971). Leaf spot incited by P. chartarum has been reported previously on several hosts worldwide (Farr & Rossman Citation2014). The hosts include lettuce (Lactuca sativa L.) (Williams & Liu Citation1976), tomato (Solanum lycopersicum L.) (Peregrine & Ahmad Citation1982), wheat (Triticum aestivum L.) (Toth et al. Citation2007), winter cherry (Withania somnifera (L.) Dunal) (Verma et al. Citation2008), switchgrass (Panicum virgatum L.) (Vu et al. Citation2013) and smooth bromegrass (Bromus inermis Leyss.) (Eken et al. Citation2006). Based on cultural, morphological and molecular characteristics, and pathogenicity to the host plant, the isolates obtained from cabbage were identified as P. chartarum. To the best of our knowledge, this is the first report of leaf spot caused by P. chartarum on cabbage in Malaysia and worldwide.
Acknowledgements
We are very thankful to the staff of Malaysian Agricultural Research and Development Institute (MARDI), and Jabatan Pertanian Malaysia for their cooperation to collect the isolates.
References
- Eken C, Jochum CC, Yuen GY. 2006. First report of leaf spot of smooth bromegrass caused by Pithomyces chartarum in Nebraska. Plant Dis. 90:108. doi:10.1094/PD-90-0108C
- Ellis MB. 1971. Dematiaceous hyphomycetes. Kew(England): CMI press.
- Farr DF, Rossman AY. 2014. Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. [Internet]; [cited 2014 Jan 15]. Available from: http://nt.ars-grin.gov/fungaldatabases/
- Food and Agriculture Organization of the United Nations. 2013. [Internet]; [cited 2013 Jun 20]. Available from: http://faostat.fao.org/
- Gonzlez-Mendoza D, Argumedo Delira R, Morales Trejo A, Pulido Herrera A, Cervantes-Daz L, Grimaldo Juarez O, Alarcn A. 2010. A rapid method for isolation of total DNA from pathogenic filamentous plant fungi. Genet Mol Res. 9:162–166. doi:10.4238/vol9-1gmr680
- Nazerian E, Sijam K, Mior Ahmad ZA, Vadamalai G. 2011. First report of cabbage soft rot caused by Pectobacterium carotovorum subsp. carotovorum in Malaysia. Plant Dis. 95:491. doi:10.1094/PDIS-09-10-0683
- Peregrine WTH, Ahmad KB. 1982. Brunei: A first annotated list of plant diseases and associated organisms. Phytopathol Pap. 27:1–87
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi:10.1093/molbev/msr121
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 22:4673–4680. doi:10.1093/nar/22.22.4673
- Toth B, Csosz M, Dijksterhuis J, Frisvad JC, Varga J. 2007. Pithomyces chartarum as a pathogen of wheat. J Plant Pathol. 89:405–408
- Verma OP, Gupta RBL, Shivpuri A. 2008. A new host for Pithomyces chartarum, the cause of a leaf spot disease on Withania somnifera. Plant Pathol. 57:385. doi:10.1111/j.1365-3059.2007.01732.x
- Vu AL, Gwinn KD, Ownley BH. 2013. First report of leaf spot on switchgrass caused by Pithomyces chartarum in the United States. Plant Dis. 97:1655. doi:10.1094/PDIS-01-13-0117-PDN
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York, NY: Academic Press; p. 315–322.
- Williams TH, Liu PSW. 1976. A host list of plant diseases in Sabah, Malaysia. Phytopathol Pap. 19:1–67