Abstract
Undescribed exotic and scientifically unknown fungal species with the potential to be pathogens are often difficult to detect on imported live plant material due to their inconspicuous nature and thus represent an important risk to Canadian forests. We have developed an early warning method based on a random sampling of asymptomatic woody live plant material imported into Canada to detect such potential alien fungal species. We received 150 asymptomatic sample lots collected by Canadian Food Inspection Agency (CFIA) inspectors. Samples were analysed by cloning and sequencing the PCR-amplified fungal nuclear ribosomal internal transcribed spacer (ITS) DNA from plant tissues to reveal fungal diversity. Out of 1845 fungal clones obtained, 267 fungal operational taxonomic units (OTUs) were identified. Using phylogenetic profiling methods, two fungal OTUs were categorized as having potential for a moderate impact on Canadian forests, 37 OTUs had a low impact and 18 OTUs could not be assessed given their low genetic similarity with other ITS sequences in GenBank. In all cases, the potential risk associated with these 57 fungal OTUs is based on (i) the fact that they can be considered unknown species to science; and (ii) they belong to orders, classes, genera and families in which pathogenic species are common. Fungal introductions with potential for a moderate impact on Canadian forests were observed at a very low frequency (0.2%) in the sampling units (clones). Only 1.3% of the CFIA samples had an OTU with a potential moderate impact on Canadian forests and 74% of the samples were free of fungal OTUs that could have any potential impact. This early warning method sheds light on the suite of exotic fungi that may enter Canada via plant material. Additionally, this method provides the tools to assess the potential risk that these fungi may post to Canadian trees and determines the magnitude of asymptomatic material that harbours fungal pathogens.
Résumé
Les espèces fongiques exotiques non-décrites et inconnues de la science ayant le potentiel d’être pathogènes représentent un risque important pour les forêts canadiennes et sont souvent difficiles à détecter sur du matériel de plantes vivantes importé à cause de leur nature cryptique. Une méthode d’alerte précoce basée sur un échantillonnage aléatoire de matériel sur des plants ligneux vivants et asymptomatiques importés au Canada a été utilisée pour détecter des champignons précédemment non décrits et étrangers à potentiel pathogène. Nous avons traité 150 lots d’échantillons asymptomatiques collectés par des inspecteurs de l’Agence canadienne d’inspection des aliments (ACIA). Les échantillons ont été analysés par clonage et séquençage des régions génomiques encodant les espaceurs internes transcrits de l’ADN ribosomal fongique (régions ITS) présents dans les tissus des plants. À partir des 1845 clones de champignons ainsi obtenus, 267 unités taxonomiques opérationnelles (UTO) fongiques ont été définies. Par une analyse de profilage phylogénétique, nous avons catégorisé deux UTO fongiques comme pouvant avoir un potentiel d’impact modéré sur les forêts canadiennes, 37 UTO avec un potentiel d’impact faible et 18 UTO restent non évaluées à cause de leur faible similarité génétique dans les banques de séquences ITS références. Dans tous les cas, le danger potentiel de ces 57 UTO fongiques provient du fait qu’elles peuvent être considérées comme de nouvelles espèces pour la science ou appartenir à un ordre, une classe, une famille ou un genre au sein duquel la présence de pathogènes est commune. L’introduction de champignons étrangers ayant un potentiel d’impact modéré pour les forêts canadiennes s’observe à une fréquence très faible (0,2%) du nombre d’unité d’échantillonnage (les clones). Seulement 1,3% des échantillons de l’ACIA ont au moins une UTO fongique avec un potentiel d’impact modéré sur les forêts canadiennes, alors que 74% des échantillons sont exempts d’UTO fongiques pouvant avoir un impact. Cette méthode d’alerte précoce fournie un portrait des communautés fongiques entrantes au Canada sur du matériel végétal vivant en mettant en évidence les champignons à potentiel pathogène, le risque qu’ils représentent pour les arbres canadiens et la magnitude de ce phénomène.
Introduction
Emerging and unknown diseases caused by invasive alien fungal species present in imported live plant material are an important threat to Canadian and North American forests (Liebhold et al. Citation2012). The International Standards for Phytosanitary Measures (ISPM) No. 15 developed by the International Plant Protection Convention (IPPC), which addresses the need to treat wood packaging and wood dunnage, is effective in the prevention of new introductions of disease-causing exotic fungi found in wood, but it does not target live plant material, such as cuttings, bare root seedlings, seeds, ornamental plants and bonsai (SIPPC Citation2005). This type of material, which falls under the Plants for Planting category, may host inconspicuous fungi, such as fungal endophytes or latent-phase pathogens (Saikkonen et al. Citation1998; Rodriguez et al. Citation2009). These fungi may not be regulated since they might not possess a scientific name, do not induce signs and symptoms, or have a well-characterized introduction pathway. In the past, the introduction of inconspicuous fungi (and Oomycota) has been responsible for diseases such as chestnut blight, white pine blister rust, scleroderris canker (European race), butternut canker and sudden oak death (Hamelin et al. Citation1998; Et-Touil et al. Citation1999; Dutech et al. Citation2010; Broders & Boland Citation2011; Goss et al. Citation2011). Due to their inconspicuous nature and the fact that these fungi were generally unknown to science before their introduction into North America, it was difficult in the 20th century for border inspectors to intercept them before their introduction and establishment into our forests. This problem is still present today.
Live plants and endophytic fungi live in an ongoing symbiotic relationship that can range from mutualistic to parasitic (Redman et al. Citation2001; Kogel et al. Citation2006; Rodriguez & Redman Citation2008). Pathogenicity is a characteristic of a given species or genus involved in an evolutionary relation with a host (Pirofski & Casadevall Citation2012). Some fungal genera may only contain saprophytic species, while other genera demonstrate a variety of symbiotic interactions ranging from mutualistic endophytes to parasitism (Redman et al. Citation2001). Pathogenic fungi that have co-evolved over a long time period with their host tend to become endophytic with that host (Saikkonen et al. Citation1998). While endophytes can be very beneficial to their host plant by providing enhanced disease resistance, herbivore deterrence and tolerance to drought and other abiotic factors (Rodriguez et al. Citation2009), they may also be latent pathogens that can become plant pathogens after host switch (Redman et al. Citation2001). Thus many endophytes can be latent pathogens and as endophytes that may exist asymptomatically, they cannot be detected by observation-based strategies. However, characterization of a fungal species generally reflects the lifestyle that the species most frequently expresses rather than the lifestyle they are capable of expressing (Rodriguez & Redman Citation1997). For example, a Colletotrichum magna isolate was found to be an endophyte on tomato, a saprophyte on dead plant matter, and a pathogen on cucumbers (Freeman & Rodriguez Citation1993). A long-time co-evolved pathogen on exotic host trees exhibiting only an endophytic lifestyle in its natural forest ecosystem abroad may become a potent pathogen once accidentally introduced on North American tree species (Magasi & Pond Citation1982). In essence, most healthy plant material moving around the world could be colonized by latent pathogens (Sakalidis et al. Citation2013). We are using these characteristics to search for latent pathogenic fungi introduced on asymptomatic imported live woody ornamental plant material with potential to become emerging forest diseases. We especially focus on endophytes belonging to genera in which forest pathogens are common. Since little is known about the fungal diversity in Canadian forests, previously undescribed or not molecularly referenced fungal species of foreign origin i.e. not yet found in North America until proven otherwise and pending further investigations, were hereafter termed alien or exotic fungal species.
In this paper, we describe the development of an early warning method based on a random sampling of asymptomatic live plant material imported into Canada and the direct nuclear ribosomal ITS DNA cloning and sequencing of their fungal inhabitants, without prior culturing on growth media, in order to detect alien forest fungal species in their endophytic stage with the potential to become emerging forest diseases. Since residence time, defined as time elapsed since introduction, is the most determining factor explaining the spread of alien fungi (Desprez-Loustau et al. Citation2010), early warning is key to control success. Additionally, this study aims to: (i) provide a portrait of the incoming fungal community acting as an indication of potential invasive alien forest fungal species and potential emerging forest diseases; and (ii) generate information on the fungal taxa found, i.e. taxonomic rank (species, genera and families), detection frequency, host species, country of origin and their potential impact on Canadian forests.
Materials and methods
Sample description
A sample lot consisting of asymptomatic dormant twigs, stems and cuttings (usually without leaves/needles) of woody plants from the same host species, same provenance (nursery or wholesaler), same country and same port of entry, was collected by Canadian Food Inspection Agency (CFIA) inspectors during routine examination of imported plant material during the 2008–2010 period. We received 150 sample lots from CFIA inspectors from the province of Quebec’s points of entry as part of a pilot study between the Canadian Forest Service and the CFIA. The large majority (141/150) originated from the USA, four came from France, three from the Netherlands and two from Thailand. American samples originated from 10 states, i.e. New York (one nursery and one sample), Oregon (18 nurseries and 92 samples), Ohio (three nurseries and seven samples), Pennsylvania (one nursery and one sample), Washington (one nursery and one sample), Minnesota (one nursery and 32 samples), California (one nursery and one sample), Vermont (one nursery and four samples), Montana (one nursery and one sample) and Florida (one nursery and one sample). To reduce analysis costs and speed up processing, the majority of sample lots were grouped by host plant genus, nursery or country of origin.
DNA extraction, amplification, cloning and sequencing
Plant material was typically dormant and consisted of a few twigs or stem cuttings that were shipped by CFIA inspectors and kept at −20ºC until needed. They were surface-sterilized (Stefani & Bérubé Citation2006) and then buds, bark (and phloem) and wood (xylem and some phloem) samples were separated and thin-sliced using a scalpel. In a few cases, conifer needles present on the samples were also processed. About 50 mg of each plant tissue were sub-sampled after grinding in a Christison M3 Mixermill (tungsten bead, 2 × 2 min at 30 hertz without extraction buffer and then 1 × 1 min at 30 hertz with extraction buffer). DNA extraction was done with the Plant DNeasy mini kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s instructions. One microlitre of the eluate was used as genomic DNA (gDNA) template for PCR. Internal transcribed spacer (ITS) regions were amplified using the fungal specific ITS1-F (Gardes & Bruns Citation1993) and the universal ITS4 (White et al. Citation1990) primers to build fungal clone libraries to reveal fungal diversity present in plant material.
The PCR mixture was made up of 10× PCR buffer, 1.6 mM MgCl2, 1.25 mM of each deoxynucleotide triphosphate, 25 μg of bovine serum albumin (BSA) (SIGMA, St. Louis, MO), 12.5 μM of each primer, and 1 unit of Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany), in a total volume of 25 μL. Thermal cycling conditions to amplify fungal DNA in plant material for the pre-cloning PCR steps to reach low stringent conditions were as follows: initial denaturation at 95°C for 2 min, 30 cycles at 94°C for 45 s, 50°C for 1 min, 72°C for 1 min 30 s, and a final elongation at 72°C for 10 min. PCR reactions were done on an MJ Research PTC-200 (MJ Research Inc., Waltham, MA). Amplified DNA concentrations were measured using a Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE). PCR products were purified with QIAquick PCR Purification Kit and cloned with QIAGEN PCR Cloning plus Kit (QIAGEN, Rockville, MD) according to the manufacturer’s instructions to reveal the fungal diversity contained in the amplified DNA. Fungal DNAs from each tissue sample from a sample lot were mixed together in equal amount to achieve a five-fold molar excess of PCR products to be incubated for 2 h at 14°C with the pdrive-cloning vector according to the manufacturer’s instructions (QIAGEN). After an overnight incubation at 37°C, 48 white bacterial colonies were spiked and transferred into the 25 μL PCR mixture for amplification. Thermal cycling conditions to amplify bacterial colonies were as follows: initial denaturation at 95°C for 2 min, 37 cycles at 94°C for 45 s, 58°C for 1 min, 72°C for 1 min 30 s and a final elongation at 72°C for 10 min.
All amplicons were sequenced on a 96-capillary 3730xl DNA analyser at the Plateforme de séquençage et de génotypage des génomes Centre de Recherche du CHUL (CHUQ) (Québec, QC, Canada). The process, from the DNA extraction of grouped sample lots to PCR amplification, cloning and sequence analysis, is called a cloning run.
Sequence analysis
To discriminate between closely related species and ensure a potentially undescribed latent phase emerging pathogen that was closely related to described ones was not missed, each clone sequence was identified with the closest sequences found in the National Center for Biotechnology Information (NCBI) GenBank database using the BLAST program (Altschul et al. Citation1990; Benson et al. Citation2008). Cloned sequences with similar BLAST results were then assembled with Sequencher version 4.6 (GeneCodes, Ann Arbor, MI). The similarity threshold for sequence clusters belonging to the same OTU was set to the high value of 98% to serve as a proxy for ‘fungal taxa’ or ‘fungal species’ in this study (O’Brien et al. Citation2005; Nilsson et al. Citation2008; Stefani et al. Citation2009). We used the 98% sequence similarity and not 95% (Arnold et al. Citation2007) as the lower level could miss a new potential emerging pathogen. Taxonomic identification of OTU was defined as follows: for sequence similarities between 98% and 92% with other ITS sequences in GenBank, the clones were considered to belong to the same genus; for sequence similarities between 92% and 85%, the clones were considered to belong to the same family and order, without distinguishing where family ends and order begins. ITS sequences from each Sequencher cluster were aligned using the ClustalW algorithm (Thompson et al. Citation1994) in MegAlign version 5.05 (DNASTAR, Madison, WI) with the default parameter settings. Alignments were edited manually with Genedoc version 2.6.002 (Nicholas et al. Citation1997). PCR-generated chimeric sequences were determined from BLAST hits displaying conspicuous incongruence between the ITS1 and ITS2 regions and were excluded from the datasets.
We tested our method with pathogen-positive samples to ascertain its capacity to detect and identify a fungus causing a forest disease. Asymptomatic portions of butternut twigs from field-collected branches infected with Sirococcus clavigignenti-juglandacearum Nair, Kostichka & Kuntz, responsible for butternut canker (Broders & Boland Citation2011), were included along with the CFIA samples.
Risk assessment and pathogenic criteria
The potential impact on Canadian forests of fungal clones representing potential undescribed alien species was evaluated by using the BLAST clustering identity with ITS sequences as described above. The following criteria were defined: (i) High potential risk category: sequence similarity above 98% permitting identification of the fungal OTU to a well-known, high-risk and well-characterized foreign forest pathogen not already present in North America, for example Chalara fraxinea (Queloz et al. Citation2011); (ii) Moderate potential risk category: unknown fungus with a sequence similarity between 90% and 98% permitting the association of the fungal OTU to a genus, sometimes family, in which virulent forest pathogens are common (Farr et al. Citation1995; Sinclair & Lyon Citation2005). These sequences were not found to match DNA sequences (>98% similarity) from North American environmental samples; (iii) Low potential risk category: unknown fungus with a sequence similarity between 85% and 98%, permitting the association of the fungal OTU to a genus or family in which moderate or low virulence pathogens are common, but with low virulence reported on woody host species (Farr et al. Citation1995; Sinclair & Lyon Citation2005); (iv) Impossible-to-evaluate impact category: unknown fungus with a sequence similarity below 85% which does not reasonably permit the association of the fungal OTU to a genus, family or order in which pathogens are present.
Phylogenetic analyses
Phylogenetic analyses were performed to better assess the lineage and the pathogenic potential of the potentially threatening fungal OTUs identified (Koski & Golding Citation2001). Homology searches were first performed using the consensus sequence of an OTU as query using the NCBI’s BLAST program (Altschul et al. Citation1990). Among the best BLAST hits obtained from GenBank, representative homologous sequences were selected using BLAST-EXPLORER to avoid duplicate species (Dereeper et al. Citation2010) (E-value cut-off: 1e-05). Additional sequences belonging to the same taxonomic lineage for each OTU were selected using the taxonomic browser from the NCBI website by focusing on reference fungal forest pathogens. The sets of sequences were aligned using MAFFT version 7.037b (default mode) (Katoh & Standley Citation2013) implemented in SeaView version 4.3.3 (Gouy et al. Citation2010). Alignments were checked and trimmed for some overhanging ends with Gblocks version 0.91b (Castresana Citation2000) implemented in SeaView using the relaxed mode (allow smaller final blocks, gap positions within the final blocks and less strict flanking positions options). Tree construction was executed using the maximum likelihood (ML) algorithm PhyML version 3.0 (Guindon & Gascuel Citation2003) implemented in SeaView and was based on the General Time Reversible model of evolution. Initial tree(s) for the heuristic search were obtained automatically using the BioNJ method (Gascuel Citation1997). To root the trees, outgroup sequences of Neocomospora africana (AF178412.1) and Discula quercina (GQ452258.1) were chosen to allow for accurate ITS sequences alignment to estimate a valid phylogeny. The bootstrap consensus tree inferred from 500 replicates was taken to represent the evolutionary history of the taxa analysed (Felsenstein Citation1985).
Results
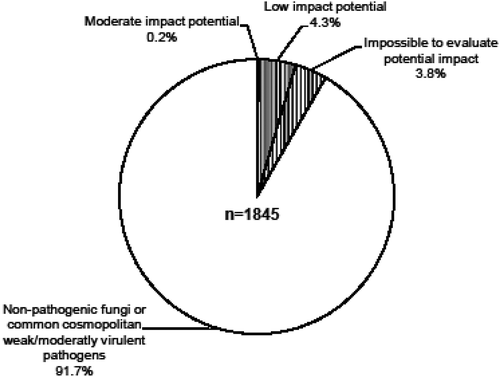
In this study, 150 sample lots from 99 exotic host plant species were extracted and amplified for their fungal nuclear ribosomal ITS DNA sequences. When there was an opportunity, some of them were grouped according to host plant genus, nursery or country of origin to reduce the number of cloning runs. Average amplified fungal DNA per tissue type was 37.1 ng/µL in the bark (and phloem), 41.7 ng/µL in the xylem (wood), 47.3 ng/µL in the needles and 57.9 ng/µL in the buds. In the end, 56 cloning runs were made (including resampling and reruns) and we obtained 1845 fungal clones. All sample lots yielded fungal clones. One or more representative nucleotide sequence of OTUs with a potential impact on Canadian forests was deposited in the NCBI GenBank database (). The 1845 fungal clones belonged to five different phyla and subphyla (Ascomycota, Basidiomycota, Ustilaginomycotina, Glomeromycota, Pucciniomycotina), 17 classes, 33 orders, 44 fungal families and 96 genera.
Table 1. Exotic fungi found on live plant material imported into Canada. Incidence of fungal clones of each OTU representing a potential risk for Canadian forests with year of collection, host, country of origin, best BLAST match, putative identification and risk assessment.
From the disease-positive samples used to test our method, the fungal pathogen S. clavigignenti-juglandacearum was identified in 69% of the clones obtained for that cloning run, indicating the capacity and efficacy of the system to intercept and identify fungal pathogens present in asymptomatic live plant tissue.
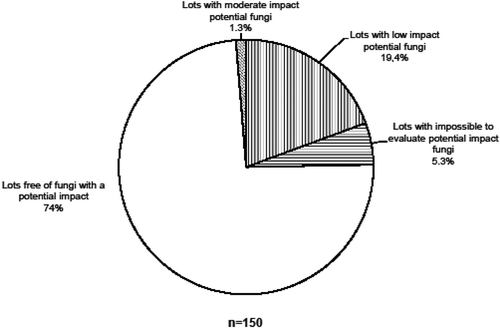
Even though the grouping of sample lots into a single cloning run blurs the direct association between a lot and a potentially threatening species, we conclude that only 1.3% of the CFIA samples had an OTU with a potential moderate impact on Canadian forests (), 19.4% of the samples had at least one OTU with a potential low impact on Canadian forests, 5.3% of the samples had at least one OTU with a potential impact that was impossible to evaluate, and 74% of the samples were free of fungal OTUs that could have a potential impact.
Fig. 1 Percentage of imported live plant material with low to moderate potential impact on Canadian forests.
The majority (94%) of the samples in this study came from the USA, which also yielded the majority of the species whose impact was low, moderate or impossible to evaluate. One mixed lot from the Netherlands and the USA provided one taxon associated with the low potential impact category.
A total of 267 OTUs referred to as fungal taxa/species in this study (groups of sequences of clones with the same putative taxonomic identification and with a sequence similarity above 98%) were identified from the 1845 clones. A total of 119 taxa belonged to a specific species, 75 were identified to the genus rank, 22 to family and 51 to the order or higher levels. The 75 taxa identified at the genus level may be known to science but not yet present in DNA databases such as GenBank. The remaining 73 taxa (22 + 51) were only identifiable at the family level (or higher) and are probably unknown species to science. These 148 (75 + 73) undescribed species were probably introduced into Canada in the last 3 years, averaging nearly 50 annual introductions, and may represent a potential threat to Canadian forests. It also highlights the contribution of this method, which bypasses the necessity to cultivate fungi on growth media and to identify species unknown to science.
The eight most common fungal species, found at a clonal frequency above 2%, were Aureobasidium pullullans (and its closely related taxonomic species which were difficult to separate using ITS sequences) at a frequency of 16.3%, Alternaria alternata (and closely related species) at 6.3%, Epicoccum nigrum at 4.2%, Botrytis cinerea at 3.7%, Cladosporium tenuissinum at 3.2%, Fusarium lateritium (and closely related species) at 2.9%, Lewia infectoria at 2.5%, and a Hormonema sp. 1 at 2.2%. Fourteen other species were found at frequencies varying between 1 and 2%. They were Phoma sojicola at a frequency of 1.9%, Cryptococcus victoriae (and closely related species) at 1.9%, Coniothyrium fuckelii at 1.8%, Phoma pinodella at 1.8%, Davidiella tassiania at 1.6%, a Dothideomycetes sp. 7 at 1.6%, Rhodotorula creatinivora at 1.5%, Botrytis fabae at 1.4%, Rossellinia nectrioides at 1.3%, Cadophora luteo-olivacea at 1.1%, Phoma herbarum at 1.1%, Allantophomopsis lycopodina at 1.1%, Phoma exigua at 1%, and a Phaeosphaeria sp. 1 at 1%. Cryptococcus victoriae and 10 other closely related Cryptococcus species were found. We did not find Cryptococcus gatti (Vanbreus & Takashio) Kwon-Chung & Boekhout, a human pathogen present in forests of British Columbia (Stephen et al. Citation2002; MacDougall et al. Citation2007).
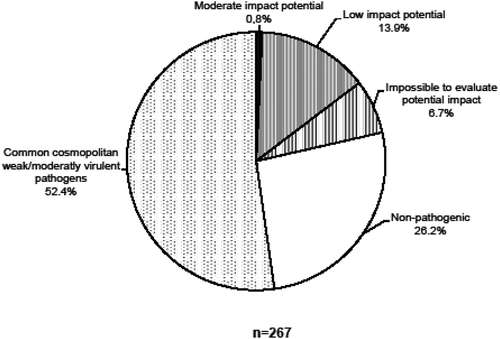
The phylogenetic clustering of a taxon within a family, genus or species based on its sequence homology was used to infer their potential to be a pathogen, its degree of virulence, and its capacity to colonize Canadian forest trees. Based on our defined criteria, two exotic fungal taxa may have a potential moderate impact on Canadian forests while 37 other taxa may have only a low impact (). Another 18 fungal species were impossible to evaluate in terms of their impact due to a low sequence similarity found in GenBank that only allowed identification to a class or phylum. Therefore, a total of 57 taxa were found to have a potential impact out of the 267 found in this study (). The 209 other fungal species were either classified as non-pathogenic (70 species) or weakly to moderately virulent pathogens that are common and cosmopolitan (139 species), including a few pathogens found on tropical hosts exclusively. A total of 21.3% (57 out of 267) of the taxa may present some degree of potential threat to Canadian forests and they were found at a frequency of 8.3% (155 out of 1845 clones). Noticeably, the number of fungal taxa representing a potential moderate impact was only two out of 267 (0.7%), with a frequency of 0.2% (4 clones out of 1845) ().
Fig. 3 Incidence and potential impact on Canadian forests of fungal clones found on imported live plant material.
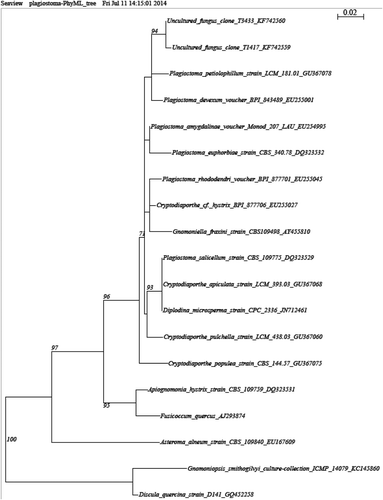
We draw attention to two taxa that are believed to have a moderate potential impact on Canadian forests. In particular, we detected a Nectriaceae sp. 1 (90% sequence similarity with Neonectria ramulariae and other Neonectria spp.) belonging to a family containing numerous devastating forest pathogens (Rossman et al. Citation1999; Halleen et al. Citation2004) (). Phylogenetic affiliation showed that the Nectriaceae sp. 1 is closely related to a large group of Neonectria and Cylindrocarpon species, again stressing its forest pathogen potential (). We also found a Plagiostoma sp. 1 (BLAST best match: 94% similarity with Diplodina microsperma, Cryptodiaporthe pulchella (syn. Plagiostoma pulchellum) and Plagiostoma euphorbia) that clusters within many Plagiostoma () and was evaluated to be in the moderate potential impact category since this genus is associated with Gnomonia that causes anthracnose, including the devastating dogwood anthracnose (Sinclair & Lyon Citation2005).
Fig. 4 Phylograms of best trees obtained by ML from ribosomal ITS sequences of the top two fungal taxa identified as having a moderate potential impact. Support values are provided only for nodes that received ≥70% ML bootstrap. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Taxa are identified next to the branch by their genus and species names, culture identification and GenBank accession number. a, Nectriaceae sp. 1, b, Plagiostoma sp. 1.
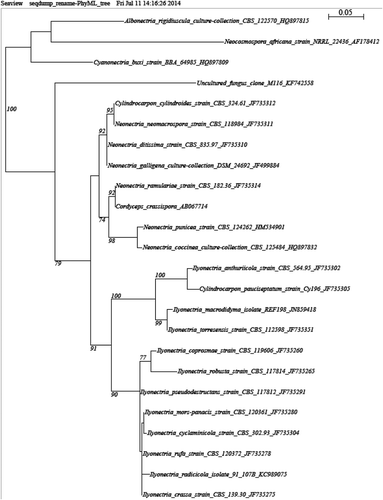
Two other taxa, a Dermateaceae sp. 1 and a Lophiostomaceae sp. 1 (), were evaluated to be in the low potential impact category. Lower sequence similarity places them both only at family level groups in which forest pathogens are present but they are all weakly virulent (Sinclair & Lyon Citation2005; Cannon & Kirk Citation2007). Five other distinct taxa of possible Coniothyrium identified as Leptosphaeriaceae sp. 4, Coniothyrium sp. 3, Coniothyrium sp. 2, Dothideaceae sp. 2 and Leptosphaeriaceae sp. 1 were also determined to be in the low potential impact category (). It is worth mentioning that 19 species from that genus are found on forest host species, but they are all weakly virulent (Przybyl Citation2002). Finally, we found a Sclerotiniaceae sp. 1, a family in which pathogens are common (Sinclair & Lyon Citation2005; Bolton et al. Citation2006; Cannon & Kirk Citation2007).
Discussion
The rDNA sequence-based method developed in this study allows the detection and identification of fungi unknown to science, thus making it possible to discover exotic asymptomatic fungi and latent pathogens with a potential to become emerging diseases. It also circumvents the limitations of DNA probe-based methods, which generally deal only with known and well-characterized organisms with available sequence data (Szemes et al. Citation2005; Van Doorn et al. Citation2009).
While no clear and immediate danger associated with species with a high impact potential, such as Ophiostoma polonica, was found in the CFIA samples processed in the present study, two fungal species represented a potential moderate threat, 37 represented a potential low-level threat and 18 were impossible to evaluate. This represents 21.4% of the species found in this study, with a frequency of 8.3% of all clones. The risk represented by these frequencies is significant even though the occurrence of every species is low (not meaningful) since 40 of these species were found only once or twice. These numbers provide a snap-shot of the incoming fungal introductions and the potential associated risk of invasive fungi that may have some degree of potential impact on Canadian forests (in at least 20.7% of the sample lots sent by the CFIA). Noticeably the species with a potential moderate impact on Canadian forests only represent 1.3% of the CFIA samples.
Most of the species found at frequencies above 1% are considered to be common cosmopolitan fungi, and while many are plant pathogens, we do not consider their introductions as adding extra risk to Canadian forests as they are already commonly found in Canada (Farr et al. Citation1995). Among the eight most common species, many are generally found on decaying plant material (Farr et al. Citation1995; Sinclair & Lyon Citation2005). They were retrieved despite the application of a surface sterilization procedure, suggesting they may have penetrated into the asymptomatic live plant samples and could be endophytes (microfungi that live within healthy plant tissues without causing any visible symptoms) (Rodriguez et al. Citation2009).
The first two species listed in (Nectriaceae sp. 1 and Plagiostoma sp. 1) can be considered threats and, even though they were found at low frequencies, the risk potential is high since a single introduction could be catastrophic (Purdy et al. Citation1985). They both belong to genera or families in which forest pathogens are common and virulent (Sinclair & Lyon Citation2005; Cannon & Kirk Citation2007). For all the other OTUs, the potential impact is based on the observation that while these fungi are unknown to science, they belong to genera and families in which pathogenic species are common. They therefore represent a threat that is hard to evaluate due to the lack of available scientific information. The large majority of the fungal species representing a level of impact that was low, moderate or impossible-to-evaluate came from the USA since almost all plant material originated from that country. The two fungal species which represent a potential moderate potential threat are already present in the USA but not reported from Canada.
Nearly 64% of the plant samples intercepted at the province of Quebec’s points of entry came from Oregon and not from nearby New York and Pennsylvania. Since Oregon’s typically cool and wet weather conditions may promote fungal sporulation and plant colonization, it is not known whether the dominance of shipments from Oregon could represent an added risk. However, samples labelled ‘American’ could be of recent foreign origin. After a year in a storage yard, or after vegetative propagation, sample origin becomes its present location and no traceability system is in place in the USA or Canada to ascertain the previous origin of ornamental plants. The same can be said of ornamental material from the Netherlands, which may originate from Eastern Europe or Asia. Thus, pathway analysis is difficult as there is no way of determining where or at what point the alien fungi colonized the plant material.
ITS sequences were recently proposed as the primary fungal barcode marker (Schoch et al. Citation2012) over other rSSU or rLSU, in part because of their relatively low level of intraspecific variation compared with their high level of interspecific variation (Nilsson et al. Citation2009; Gazis et al. Citation2011). Hence, the method described here only allows one to arbitrarily assign a pathogenic potential to an alien fungal species by comparing its lineage with well-known pathogenic fungal species without determining its real pathogenic characters, such as invasiveness and virulence on a new plant host. This is clearly a weakness of the method as it is based on associations. We are aware that our capacity to evaluate risk is limited by the use of ribosomal ITS phylogenetic analysis. For example, an undescribed fungus found in an ITS gene phylogenetic analysis cluster within the Mycosphaerella genus is an indicator of some potential risk, but of limited value since there are possibly thousands of species in that genus, many of which are not pathogenic (Crous et al. Citation2007) and there is little sequence variation to differentiate them. There is also the possibility that a new virulent pathogen may arise in a genus not known to cause disease as we have seen with Geosmithia morbida, the causal agent of Thousand cankers disease in walnut (Kolarik et al. Citation2011). Until we have more data on fungal genes involved in pathogenicity and virulence, phylogenetic analysis of ITS rDNA regions in environmental samples remains our best tool to evaluate the potential risk of a given OTU in the absence of live isolates and in vivo host range tests. Other molecular detection methods using ITS rDNA region or coding genes are available (Hamelin Citation2006); however, they usually target specific pathogens and not whole fungal communities simultaneously (Filion et al. Citation2004).
Since the sequence length can interfere with the produced score of a BLAST search, the closest sequence match is not always the best choice. Thus, a phylogenetic analysis remains a better way to ascertain the identity of an unknown species. The phylogenetic analysis used to evaluate potentially threatening OTUs is based on ITS sequence loci, which have no linkage to pathogenicity genes. ITS loci can produce phylogenetic trees lacking statistically significant support at the family or higher taxonomic levels (Jeffroy et al. Citation2006) due to the limited phylogenetic information present in that region. Nevertheless, it remains our best tool to assign a potential risk level considering the limited number of commonly available fungal DNA markers in databases (Rossman Citation2007; Chase & Fay Citation2009; Schoch et al. Citation2012). Future multiloci sequencing will be a valuable tool to boost phylogenetic risk evaluation based on multi-DNA markers since ribosomal genes do not always discriminate between closely related species, potentially hiding cryptic invasive fungal species with a potential to become an emerging pathogen (Gazis et al. Citation2011).
The number of samples in this pilot project is insufficient to create a system capable of intercepting a significant portion of the fungi introduced into Canada. A sample size one order of magnitude greater than the one used in this report would begin to provide a statistically significant evaluation of the situation. We hope to include CFIA samples from the port of Vancouver (Canada), an important port of entry for invasive pests, and future efforts will target samples from that area. Urban trees and urban forests usually are the first to be affected by exotic fungal species due to their proximity to ports of entry, transport hubs and plant retailers (Colunga-Garcia et al. Citation2010). Urban areas are also where exotic ornamentals are found in greatest numbers. Urban trees are especially affected by alien pests and can be the source of pest invasions in adjacent forest areas (Alston & Richardson Citation2006). They will be the focus of our future research on potential emerging forest diseases.
Fungal invasion will most likely increase along with increasing global trade (Colunga-Garcia et al. Citation2010). The fact that fungi are well-suited for long-distance dispersal, that they only need to be introduced in low numbers of propagules, and that they possess a high potential for evolutionary change makes them successful potential invaders (Purdy et al. Citation1985; Parker & Gilbert Citation2004; Desprez-Lousteau et al. Citation2007). Natural ecosystems may act as a repository for plant pathogens from which naturally occurring endophytes may have a great opportunity to expand their host range once introduced abroad and change their lifestyle, resulting in the disruption of existing co-evolved plant-fungus associations to become emerging diseases (Rodriguez & Redman Citation1997, Citation2008). The versatile lifestyle potential of endophytes is directly dependent on the interaction between plant, fungi and environmental conditions (Schulz & Boyle Citation2005). Lack of co-evolution and specific resistance development in native plant species against alien endophytes may represent a threat by increasing the susceptibility of plants to exotic fungi. Fortunately fungal infection is the exception rather than the rule due to the broadly accepted principle called non-host resistance. Fungi face constitutive defences and induced defence mechanism that result in general plant immunity (Uma et al. Citation2011). However, endemic fungal species with potential virulence are rarely detected, perhaps hidden as endophytes and latent pathogens. A high frequency of colonization of internal and healthy plant tissues may attest to their low or absence of virulence on the carrier host plant (Sieber Citation2007) but is not reflective of their power of invasiveness, which still remains little known and studied.
Acknowledgements
The first author thanks Julie Dubé (Canadian Forest Service – Laurentian Forestry Centre) for her laboratory work, CFIA inspectors for collecting and shipping the plant samples, Baptiste Petiot and Aurore Lenglet for their work during their internship at our laboratory, and Jean-Guy Champagne and Louis-Philippe Vaillancourt from CFIA for making this study possible.
Additional information
Funding
References
- Alston KP, Richardson DM. 2006. The roles of habitat features, disturbance, and distance from putative source populations in structuring alien plant invasions at the urban/wildland interface on the Cape Peninsula, South Africa. Biol Conserv. 132:183–198. doi:10.1016/j.biocon.2006.03.023
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. doi:10.1016/S0022-2836(05)80360-2
- Arnold AE, Henk DA, Eells RL, Lutzoni F, Vilgalys R. 2007. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia. 99:185–206. doi:10.3852/mycologia.99.2.185
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2008. GenBank. Nucleic Acids Res. 36:D25–D30. doi:10.1093/nar/gkm929
- Bolton MD, Thomma BP, Nelson BD. 2006. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol. 7:1–16. doi:10.1111/j.1364-3703.2005.00316.x
- Broders KD, Boland GJ. 2011. Reclassification of the butternut canker fungus, Sirococcus clavigignenti-juglandacearum, into the genus Ophiognomonia. Fungal Biol. 115:70–79. doi:10.1016/j.funbio.2010.10.007
- Cannon PF, Kirk PM. 2007. Fungal families of the world. Cambridge, MA: CAB International; p. 456 pages.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. doi:10.1093/oxfordjournals.molbev.a026334
- Chase MW, Fay MF. 2009. Ecology. Barcoding of Plants and Fungi. Science. 325:682–683.
- Colunga-Garcia M, Magarey RA, Haack RA, Gage SH, Qi J. 2010. Enhancing early detection of exotic pests in agricultural and forest ecosystems using an urban-gradient framework. Ecol Applic. 20:303–310. doi:10.1890/09-0193.1
- Crous PW, Braun U, Groenewald JZ. 2007. Mycosphaerella is polyphyletic. Stud Mycol. 58:1–32. doi:10.3114/sim.2007.58.01
- Dereeper A, Audic S, Claverie J-M, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 10:8. doi:10.1186/1471-2148-10-8
- Desprez-Loustau M-L, Courtecuisse R, Robin C, Husson C, Moreau P-A, Blancard D, Selosse M-A, Lung-Escarmant B, Piou D, Sache I. 2010. Species diversity and drivers of spread of alien fungi (sensu lato) in Europe with a particular focus on France. Biol Invasions. 12:157–172. doi:10.1007/s10530-009-9439-y
- Desprez-Loustau M-L, Robin C, Buée M, Courtecuisse R, Garbaye J, Suffert F, Sache I, Rizzo DM. 2007. The fungal dimension of biological invasions. Trends Ecol Evol. 22:472–480. doi:10.1016/j.tree.2007.04.005
- Dutech C, Fabreguettes O, Capdevielle X, Robin C. 2010. Multiple introductions of divergent genetic lineages in an invasive fungal pathogen, Cryphonectria parasitica, in France. Heredity. 105:220–228. doi:10.1038/hdy.2009.164
- Et-Touil K, Bernier L, Beaulieu J, Bérubé JA, Hopkin A, Hamelin RC. 1999. Genetic structure of Cronartium ribicola populations in eastern Canada. Phytopathology. 89:915–919. doi:10.1094/PHYTO.1999.89.10.915
- Farr DF, Bills GF, Chamuris GP, Rossman AY. 1995. Fungi on plants and plant products in the United States. Second Edition ed. St. Paul, MN, USA: APS Press; p. 1252 pages.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791. doi:10.2307/2408678
- Filion M, Hamelin RC, Bernier L, St-Arnaud M. 2004. Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl Environ Microbiol. 70:3541–3551. doi:10.1128/AEM.70.6.3541-3551.2004
- Freeman S, Rodriguez RJ. 1993. Genetic conversion of a fungal plant pathogen to a nonpathogenic, endophytic mutualist. Science. 260:75–78. doi:10.1126/science.260.5104.75
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 2:113–118. doi:10.1111/j.1365-294X.1993.tb00005.x
- Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 14:685–695. doi:10.1093/oxfordjournals.molbev.a025808
- Gazis R, Rehner S, Chaverri P. 2011. Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Mol Ecol. 20:3001–3013. doi:10.1111/j.1365-294X.2011.05110.x
- Goss EM, Larsen M, Vercauteren A, Werres S, Heungens K, Grünwald NJ. 2011. Phytophthora ramorum in Canada: evidence for migration within North America and from Europe. Phytopathology. 101:166–171. doi:10.1094/PHYTO-05-10-0133
- Gouy M, Guindon S, Gascuel O. 2010. SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. doi:10.1093/molbev/msp259
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. System Biol. 52:696–704. doi:10.1080/10635150390235520
- Halleen F, Schroers H-J, Groenewald JZ, Crous PW. 2004. Novel species of Cylindrocarpon (Neonectria) and Campylocarpon gen. nov. associated with black foot disease of grapevines (Vitis spp.). Stud Mycol. 50:431–455.
- Hamelin RC. 2006. Molecular epidemiology of forest pathogens: from genes to landscape. Can J Plant Pathol. 28:167–181. doi:10.1080/07060660609507285
- Hamelin RC, Lecours N, Laflamme G. 1998. Molecular evidence of distinct introductions of the European race of Gremmeniella abietina into North America. Phytopathology. 88:582–588. doi:10.1094/PHYTO.1998.88.6.582
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22:225–231. doi:10.1016/j.tig.2006.02.003
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. doi:10.1093/molbev/mst010
- Kogel K-H, Franken P, Hückelhoven R. 2006. Endophyte or parasite – what decides? Curr Opin Plant Biol. 9:358–363. doi:10.1016/j.pbi.2006.05.001
- Kolarik M, Freeland E, Utley C, Tisserat N. 2011. Geosmithia morbida sp. nov., a new phytopathogenic species living in symbiosis with the walnut twig beetle (Pityophthorus juglandis) on Juglans in USA. Mycologia. 103:325–332. doi:10.3852/10-124
- Koski LB, Golding GB. 2001. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 52:540–542. doi:10.1007/s002390010184
- Liebhold AM, Brockerhoff EG, Garrett LJ, Parke JL, Britton KO. 2012. Live plant imports: the major pathway for forest insect and pathogen invasions of the US. Front Ecol Environ. 10:135–143. doi:10.1890/110198
- MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, Kronstad JW, Morshed MG, Bartlett KH. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 13:42–50. doi:10.3201/eid1301.060827
- Magasi LP, Pond SE. 1982. European larch canker: a new disease in Canada and a new North American host record. Plant Dis. 66:339–340. doi:10.1094/PD-66-339
- Nicholas KB, Nicholas Jr HB, Deerfield DW. 1997. GeneDoc: analysis and visualization of genetic variation. Embnews News. 4:14.
- Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson K-H. 2008. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinf. 4:193–201.
- Nilsson RH, Ryberg M, Abarenkov K, Sjökvist E, Kristiansson E. 2009. The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol Lett. 296:97–101. doi:10.1111/j.1574-6968.2009.01618.x
- O’Brien HE, Parrent JL, Jackson JA, Moncalvo J-M, Vilgalys R. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 71:5544–5550. doi:10.1128/AEM.71.9.5544-5550.2005
- Parker IM, Gilbert GS. 2004. The evolutionary ecology of novel plant-pathogen interactions. Annu Rev Ecol Evol Syst. 35:675–700. doi:10.1146/annurev.ecolsys.34.011802.132339
- Pirofski L-A, Casadevall A. 2012. Q&A: what is a pathogen? A question that begs the point. BMC Biol. 10:6.
- Przybyl K. 2002. Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. For Pathol. 32:387–394. doi:10.1046/j.1439-0329.2002.00301.x
- Purdy LH, Krupa SV, Dean JL. 1985. Introduction of sugarcane rust into the Americas and its spread to Florida. Plant Dis. 69:689–693. doi:10.1094/PD-69-689
- Queloz V, Grünig CR, Berndt R, Kowalski T, Sieber TN, Holdenrieder O. 2011. Cryptic speciation in Hymenoscyphus albidus. For Pathol. 41:133–142. doi:10.1111/j.1439-0329.2010.00645.x
- Redman RS, Dunigan D, Rodriguez RJ. 2001. Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader? New Phytol. 151:705–716. doi:10.1046/j.0028-646x.2001.00210.x
- Rodriguez R, Redman R. 2008. More than 400 million years of evolution and some plants still can’t make it on their own: plant stress tolerance via fungal symbiosis. J Exp Bot. 59:1109–1114. doi:10.1093/jxb/erm342
- Rodriguez RJ, Redman RS. 1997. Fungal life-styles and ecosystem dynamics: biological aspects of plant pathogens, plant endophytes and saprophytes. Adv Bot Res. 24:169–193. doi:10.1016/S0065-2296(08)60073-7
- Rodriguez RJ, White Jr Jr JF, Arnold AE, Redman RS. 2009. Fungal endophytes: diversity and functional roles. New Phytol. 182:314–330. doi:10.1111/j.1469-8137.2009.02773.x
- Rossman A. 2007. Report of the planning workshop for all fungi DNA barcoding. Inoculum. 58:1–5.
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R. 1999. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol. 42:1–248.
- Saikkonen K, Faeth SH, Helander M, Sullivan TJ. 1998. Fungal endophytes: a continuum of interactions with host plants. Annu Rev Ecol Syst. 29:319–343. doi:10.1146/annurev.ecolsys.29.1.319
- Sakalidis ML, Slippers B, Wingfield BD, Hardy GESJ, Burgess TI. 2013. The challenge of understanding the origin, pathways and extent of fungal invasions: global populations of the Neofusicoccum parvum–N. ribis species complex. Diversity Distrib. 19:873–883. doi:10.1111/ddi.12030
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci. USA. 109:6241–6246. Fungal Barcoding Consortium.
- Schulz B, Boyle C. 2005. The endophytic continuum. Mycol Res. 109:661–686. doi:10.1017/S095375620500273X
- SIPPC. 2005. International Standards for Phytosanitary Measures 1 to 24. Rome, Italy: Food and Agriculture Organization of the United Nations. https://www.ippc.int/publications/regulation-wood-packaging-material-international-trade-0
- Sieber TN. 2007. Endophytic fungi in forest trees: are they mutualists? Fungal Biol Rev. 21:75–89. doi:10.1016/j.fbr.2007.05.004
- Sinclair WA, Lyon HH. 2005. Diseases of trees and shrubs. 2nd ed.. Ithaca, NY: Cornell University Press; p. 660.
- Stefani FOP, Bérubé JA. 2006. Biodiversity of foliar fungal endophytes in white spruce (Picea glauca) from southern Québec. Can J Bot. 84:777–790. doi:10.1139/b06-022
- Stefani FOP, Moncalvo J-M, Seguin A, Berube JA, Hamelin RC. 2009. Impact of an 8-Year-Old Transgenic Poplar Plantation on the Ectomycorrhizal Fungal Community. Appl Environ Microbiol. 75:7527–7536. doi:10.1128/AEM.01120-09
- Stephen C, Lester S, Black W, Fyfe M, Raverty S. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 43:792–794.
- Szemes M, Bonants P, De Weerdt M, Baner J, Landegren U, Schoen CD. 2005. Diagnostic application of padlock probes–multiplex detection of plant pathogens using universal microarrays. Nucleic Acids Res. 33:e70. doi:10.1093/nar/gni069
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. doi:10.1093/nar/22.22.4673
- Uma B, Swaroopa Rani T, Podile AR. 2011. Warriors at the gate that never sleep: non-host resistance in plants. J Plant Physiol. 168:2141–2152. doi:10.1016/j.jplph.2011.09.005
- Van Doorn R, Slawiak M, Szemes M, Dullemans AM, Bonants P, Kowalchuk GA, Schoen CD. 2009. Robust detection and identification of multiple oomycetes and fungi in environmental samples by using a novel cleavable padlock probe-based ligation detection assay. Appl Environ Microbiol. 75:4185–4193. doi:10.1128/AEM.00071-09
- White TJ, Bruns TD, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, Eds. PCR protocols: a guide to methods and applications. San Diego: Academic Press; p. 315–322.