Abstract
Grapevine red blotch-associated virus (GRBaV) is a recently discovered ssDNA virus that is widespread in wine grapes in California. We investigated whether GRBaV infection was present in 156 table grape accessions of Vitis vinifera that included 53 accessions exhibiting leafroll-like symptoms and 81 accessions from diverse geographic origins. Cane samples were collected during the dormant season in 2012 and analysed for GRBaV infection by PCR. A total of 73 accessions showed presence of GRBaV and these included raisin and table grape accessions with black, green and red berries. A 557 bp amplicon obtained by PCR was purified and sequenced, and the phylogenetic relationship among GRBaV isolates was examined by the maximum likelihood method. The maximum genetic variability among the isolates was only 8% and they belonged to two clades. Although it is not yet known if GRBaV is present outside of North America, 54 accessions from sources originating outside of North America tested positive for the virus.
Résumé
Le virus associé à la tache rouge de la vigne (VaTRV) est un virus à ADN simple brin récemment découvert et qui est très répandu chez les raisins de cuve californiens. Nous avons étudié l’infection causée par le VaTRV afin de voir si nous la trouvions dans les 156 accessions de raisins de table Vitis vinifera, qui incluaient 53 accessions affichant des symptômes semblables à l’enroulement et 81 accessions d’origines géographiques diverses. En 2012, des échantillons de tiges ont été collectés durant la saison de dormance et analysés par PCR afin d’y déceler l’infection causée par le VaTRV. En tout, 73 accessions étaient porteuses du VaTRV et celles-ci comportaient des accessions de raisins secs et de raisins de table à fruits noirs, verts et rouges. Un amplicon de 557 bp obtenu par PCR a été purifié et séquencé, et la relation phylogénétique qui existe entre les isolats de VaTRV a été examinée par la méthode de vraisemblance maximale. La variabilité génétique maximale chez les isolats n’était que de 8 % et ceux-ci appartenaient à deux variantes. Bien que nous ne sachions pas encore si le VaTRV existe ailleurs qu’en Amérique du Nord, 54 accessions de sources provenant de régions autres que nord-américaines se sont révélées positives en ce qui a trait au virus.
Introduction
There are at least 65 viruses known to infect grapevines (Vitis vinifera L.) (Martelli Citation2014) and several of these have been found infecting grapevines in the USA. While all grapevine-infecting viruses are important from a regulatory perspective in the international movement of grapevine planting stock, Grapevine fanleaf virus and Tomato ringspot virus (Family: Secoviridae), Grapevine leafroll-associated virus −1 (GLRaV-1), GLRaV-2, GLRaV-3 and GLRaV-4 (Family: Closteroviridae), and Grapevine virus A (GVA) and GVB (Family: Betaflexiviridae) are important as they affect grapevine health and productivity in California. Prior to 2009, most of the grapevine viruses were discovered by conventional methods such as virion purification and/or dsRNA characterization followed by cDNA cloning. Since 2009, discovery of new viruses in grapevines has been accelerated by metagenomic analysis using next-generation sequencing (Al Rwahnih et al. Citation2009, Citation2012b, Citation2013; Zhang et al. Citation2011; Espach et al. Citation2012). The discovery of a new virus is generally followed by the development of appropriate molecular diagnostic assays such as ELISA, RT-PCR and PCR, for post-entry quarantine detection protocols at importing locations (Rowhani et al. Citation2005).
In Napa Valley, California, a new disease characterized by red blotches along leaf margins and within the leaf blade, and red veins noticeable on the undersurface of leaves, was observed in red grape varieties in 2008 (Al Rwahnih et al. Citation2013). A new DNA virus, Grapevine red blotch-associated virus (GRBaV), first described as Grapevine Cabernet franc-associated virus in New York (Krenz et al. Citation2012) was detected in grapevines showing red blotch symptoms (Al Rwahnih et al. Citation2013) and is a tentative member of the family Geminivirideae. In 2013, in Washington State vineyards, a disease similar to red blotch disease was reported, and a virus named Grapevine red leaf-associated virus, which is genetically identical to GRBaV, was detected (Poojari et al. Citation2013). In experimental conditions, this virus has been reported to be transmitted by the Virginia creeper leafhopper (Erythroneura ziczac Walsh; Family Erythroneurideae) to grapevines (Poojari et al. Citation2013).
The National Clonal Germplasm Repository (NCGR) for Mediterranean tree fruits, nut crops and grapes is located near Winters, CA. This collection is managed by the United States Department of Agriculture, Agricultural Research Service, and curates a repository of grapevine cultivars, breeders’ lines, and wild relatives belonging to the genus Vitis (Family Vitaceae) with diverse geographic origins. The collection contains 3381 grape accessions, representing 59 taxa from 55 countries. The mission of the NCGR is to acquire, maintain, evaluate and distribute the germplasm. These genetic resources are made available to public and private plant researchers and growers worldwide. Several viruses have been reported in grapevines at this repository (Osman et al. Citation2007, Citation2013; Al Rwahnih et al. Citation2012b).
To ascertain if NCGR accessions are infected by GRBaV, we conducted a survey of about 13% of Vitis vinifera accessions and found the virus to be prevalent.
Materials and methods
Collection of plant material
Dormant canes were collected from 153 table grape accessions at the NCGR vineyard at Winters, CA in the autumn of 2012 that included eight Vitis hybrids and the rest were V. vinifera grafted on rootstock ‘St. George’. The selected accessions represent 12.75% of the table grape accessions and included 53 accessions bearing fruit coloured black to red and exhibited leafroll-like symptoms. The rest were selected based on country of origin to provide a broad international representation.
Nucleic acid extraction and detection of GRBaV by polymerase chain reaction
Nucleic acid (NA) extracts were prepared from each of the grapevine samples as described by Al Rwahnih et al. (Citation2013). About 0.2 g of frozen bark scrapings was homogenized using a HOMEX grinder (www.bioreba.com) and NA extracts were prepared using a MagMAX™-96 viral RNA isolation kit (www.invitrogen.com) as per manufacturer’s protocol. Extracted NA samples were analysed for the presence of GRBaV by polymerase chain reaction (PCR) using primers GVGF1 and GVGR1 that flank the V2 gene, according to Al Rwahnih et al. (Citation2013). The amplified PCR products were analysed by electrophoresis using 1% agarose gel and Tris-acetate-EDTA (TAE) buffer. Amplicons of GRBaV were eluted from gels using the ZymoClean Gel DNA Recovery Kit (Zymo Research Corp, USA), quantified and sequenced using GVGF1 and GVGR1 primers by Sanger sequencing at the UC-Davis sequencing facility (http://dnaseq.ucdavis.edu).
Phylogenetic analysis of the GRBaV amplicons
Nucleotide sequences of the amplicons were aligned along with GRBaV sequences available in GenBank using Sequence Analysis and Molecular Biology Data Management software Vector NTI Advance™ 11.5 (Invitrogen, USA). Alignments were made with the default options of multiple sequence alignment program Clustal X 1.8 (Thompson et al. Citation1997) and phylogenetic analyses were conducted by the maximum likelihood method of Molecular Evolutionary Genetic Analysis software MEGA version 5.2 (Tamura et al. Citation2011). Sequences of V2 genes of GRBaV isolates in the public domain database were included in the phylogenetic analysis and V2 gene of Maize streak virus (GenBank: HQ693391) was used as an outgroup. Support for the tree nodes was estimated using 1000 bootstrap replicates with default parameters; branches corresponding to partitions reproduced in less than 70% bootstrap replicates were collapsed.
Results and discussion
Of the 156 accessions of V. vinifera analysed by PCR, 73 accessions were found to be infected by GRBaV (). The accessions that tested positive were from 33 countries and five continents. Several accessions obtained from major grape producing countries such as France, Italy, Spain and the USA, tested positive for the virus and a majority of these were also infected by one or more grapevine viruses ().
Table 1. List of table grape (Vitis vinifera L.) accessions at the National Clonal Germplasm Repository at Winters, CA, USA, infected by Grapevine red blotch-associated virus and other grapevine viruses.
When GRBaV was discovered initially, it was found only in red varieties of wine grapes (Al Rwahnih et al. Citation2013). In the NCGR accessions, grapevine cultivars grown for producing juice, raisins and table grapes were found to be infected by GRBaV. Likewise, the virus was detected in grapevines bearing black, red and green berries (). In red fruit accessions that exhibited leafroll-like symptoms, GRBaV incidence was very high (17/19).
When the amplified GRBaV products (n = 67) were sequenced and compared with those of publicly available sequences, the identity scores among them ranged from 92% to 100%. The lowest identity score found in this study was not different from the 92% identity observed between the complete genome sequences of two isolates of GRBaV CF214-1 from ‘Cabernet franc’ and isolate CS337-1 from ‘Cabernet Sauvignon’ clone 337 (Al Rwahanih et al. Citation2013). This variability is lower than that generally found in RNA viruses which is up to 24% in GLRaV-3 (Chooi et al. Citation2013), but in line with what has been seen for monopartite geminiviruses belonging to the genus Mastrevirus (Muhire et al. Citation2013). Phylogenetically, the 67 isolates were grouped under two clades () much like what was reported recently for wine grape isolates of GRBaV (Krenz et al. Citation2014). Interestingly, a vast majority of the isolates belonged to clade 2.
Fig. 1 Phylogenetic tree constructed based on the maximum likelihood method using the nucleotide sequence of V2 open reading frame of Grapevine red blotch-associated virus isolates present in 67 Vitis vinifera accessions. The tree was constructed by using the V2 gene sequence of Maize streak virus (MSV) as an outgroup. The numbers below the branch represent percent support for the clade.
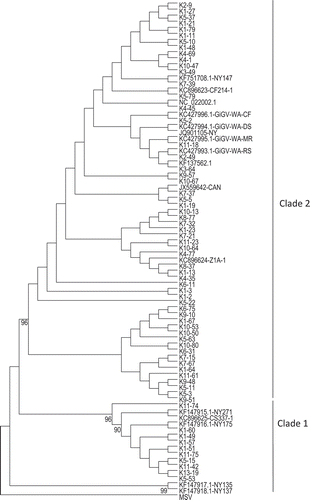
GRBaV has been discovered recently (Krenz et al. Citation2012; Al Rwahnih et al. Citation2013; Poojari et al. Citation2013); consequently not much is known about its occurrence in other major grape-producing countries outside of the USA. Also, despite implicating E. ziczac as a vector of GRBaV under laboratory conditions (Poojari et al. Citation2013), it is not known how plant to plant spread of this virus occurs under field conditions. Using the available data, we attempted to examine if genetically identical isolates were found associated together in the field. As can be inferred from , the isolates within a clade were not always from vines that were in close spatial proximity in the NCGR vineyard block and this pattern failed to support vector-mediated spread within the vineyard. Likewise, GRBaV isolates from accessions planted in close proximity in the vineyard were in different clades. For example, the virus isolate in cultivars ‘Madeleine’ (K1-49) and ‘Sovereign Rose’ ((K2-49) from Canada that are in adjacent rows are in separate clades. Also, the majority of GRBaV-infected accessions originating from a single country had isolates from the two clades. For example, isolates in accessions from ‘Duc de Malakoff’ (K5-2), ‘Estellat’ (K5-11), ‘Luquaci Thompson’ (K5-63), ‘Milton’ (K5-79), ‘Olivette Noire’ (K9-51), ‘Olivette Rose’ (K6-31) and ‘Servant’ (K1-23) from France belonged to clade 1, while the isolates in accessions ‘Oeillade Noire’ (K11-75) and ‘Poet Matabon’ (K1-60) belonged to clade 2. Many accessions were found to be co-infected with GRBaV and one or more grapevine viruses that caused leafroll and/or other diseases.
The NCGR germplasm repository provides access to genetically diverse cultivars of Vitis and their wild species originating from different geographic regions. While many are selected for introgression of superior agronomic and horticultural characteristics, they also serve as gene pools for resistance to plant pathogens and insect pests. However, it remains to be seen if any of the accessions at the NCGR would be a potential source of resistance to GRBaV. More extended testing of grapevine accessions and screening is required to identify grapevine cultivars with resistance to GRBaV which will be useful in a breeding programme.
This study represents the first report of GRBaV infection of table grapes. With the recent findings of Krenz et al. (Citation2014), it is now certain that both table and wine grapes are susceptible to GRBaV infection. Future studies should address the impact of virus infection on the quality and productivity of table grapes.
Disclaimer
Mention of a trademark, proprietary products, or vendor does not constitute guarantee or warranty of the product by the US Department of Agriculture, and does not imply its approval to the exclusion of other products and vendors that might also be suitable.
Acknowledgements
The authors acknowledge Bernard Prins, Horticulturist, NCGR, USDA-ARS, for his help in providing a list of Vitis cultivars and assistance in collecting the samples. This work was supported by USDA-ARS CRIS 5306-22000-014-00D.
References
- Al Rwahnih M, Daubert S, Golino D, Rowhani A. 2009. Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology. 387:395–401. doi:10.1016/j.virol.2009.02.028
- Al Rwahnih M, Dave A, Anderson MM, Rowhani A, Uyemoto JK, Sudarshana MR. 2013. Association of a DNA virus with grapevines affected by red blotch disease in California. Phytopathology. 103:1069–1076. doi:10.1094/PHYTO-10-12-0253-R
- Al Rwahnih M, Osman F, Sudarshana M, Uyemoto J, Minafra A, Saldarelli P, Martelli G, Rowhani A. 2012a. Detection of Grapevine leafroll-associated virus 7 using real time qRT-PCR and conventional RT PCR. J Virol Methods. 179:383–389. doi:10.1016/j.jviromet.2011.11.026
- Al Rwahnih M, Sudarshana MR, Uyemoto JK, Rowhani A. 2012b. Complete genome sequence of a novel vitivirus isolated from grapevine. J Virol. 86:9545. doi:10.1128/JVI.01444-12
- Chooi KM, Cohen D, Pearson MN. 2013. Molecular characterisation of two divergent variants of grapevine leafroll-associated virus 3 in New Zealand. Arch Virol. 158:1597–1602. doi:10.1007/s00705-013-1631-9
- Espach Y, Maree HJ, Burger JT. 2012. Complete genome of a novel endornavirus assembled from next-generation sequence data. J Virol. 86:13142. doi:10.1128/JVI.02538-12
- Krenz B, Thompson JR, Fuchs M, Perry KL. 2012. Complete genome sequence of a new circular DNA virus from grapevine. J Virol. 86:7715. doi:10.1128/JVI.00943-12
- Krenz B, Thompson JR, McLane H, Fuchs M, Perry KL. 2014. Grapevine red blotch-associated virus is widespread in the United States. Phytopathology. 104:1232–1240. doi:10.1094/PHYTO-02-14-0053-R
- Martelli GP. 2014. Grapevine-infecting viruses. J Plant Pathol. 96:136.
- Muhire B, Martin DP, Brown JK, Navas-Castillo J, Moriones E, Zerbini FM, Rivera-Bustamante R, Malathi VG, Briddon RW, Varsani A. 2013. A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol. 158:1411–1424. doi:10.1007/s00705-012-1601-7
- Osman F, Hodzicb E, Omanska-Klusekb A, Olinekab T, Rowhani A. 2013. Development and validation of a multiplex quantitative PCR assay for the rapid detection of Grapevine virus A, B and D. J Virol Methods. 194:138–145. doi:10.1016/j.jviromet.2013.07.046
- Osman F, Leutenegger C, Golino D, Rowhani A. 2007. Real-time RT-PCR (TaqMan®) assays for the detection of Grapevine leafroll associated viruses 1-5 and 9. J Virol Methods. 141:22–29. doi:10.1016/j.jviromet.2006.11.035
- Poojari S, Alabi OJ, Fofanov VY, Naidu RA. 2013. A leafhopper-transmissible DNA virus with novel evolutionary lineage in the family Geminiviridae implicated in grapevine redleaf disease by next-generation sequencing. PLoS One. 8:e64194. doi:10.1371/journal.pone.0064194
- Rowhani A, Uyemoto JK, Golino DA, Martelli GP. 2005. Pathogen testing and certification of Vitis and Prunus species. Annu Rev Phytopathol. 43:261–278. doi:10.1146/annurev.phyto.43.040204.135919
- Sánchez-Campos S, Martínez-Ayala A, Márquez-Martín B, Aragón-Caballero L, Navas-Castillo J, Moriones E. 2013. Fulfilling Koch’s postulates confirms the monopartite nature of tomato leaf deformation virus: a begomovirus native to the New World. Virus Res. 173:286–293. doi:10.1016/j.virusres.2013.02.002
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi:10.1093/molbev/msr121
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 25:4876–4882. doi:10.1093/nar/25.24.4876
- Zhang Y, Singh K, Kaur R, Qiu W. 2011. Association of a novel DNA virus with the grapevine vein clearing and vine decline syndrome. Phytopathology. 101:1081–1090. doi:10.1094/PHYTO-02-11-0034
