Abstract
Canola (Brassica napus) plants with Aster yellows-like symptoms were collected in 2011 and 2012 from canola fields in North Dakota, USA. The presence of phytoplasmas in the symptomatic plants was confirmed by a nested PCR assay using universal primer pairs P1/P7 and R16F2n/R2. Phytoplasmas were identified as ‘Candidatus phytoplasma asteris’ related strains with >99% nucleotide identity to the reference strain M30790 based on in silico analysis of the F2nR2 fragments using the iPhyClassifier online tool. Based on sequence homology searches, RFLP and phylogenetic analyses of 16S rDNA gene, phytoplasmas belonging to two distinct lineages, 16SrI-A and 16SrI-B subgroups, were identified infecting canola in North Dakota. Further characterization of the phytoplasmas based on ribosomal protein operon and secY genes also supported the presence of two distinct lineages. Sequence comparison and RFLP analyses based on the three regions confirmed the presence of mixed infection by phytoplasmas belonging to subgroups 16SrI-A and 16Sr1-B in two samples, one each in 2011 and 2012. To our knowledge, this is the first report of 16SrI-A subgroup phytoplasma and mixed infection by 16SrI-A and 16SrI-B phytoplasmas infecting canola in the USA.
Résumé
Des plants de canola (Brassica napus), affichant des symptômes semblables à ceux de la jaunisse de l’aster, ont été collectés en 2011 et 2012 dans des champs du Dakota du Nord, aux États-Unis. La présence de phytoplasmes dans les plants symptomatiques a été confirmée par PCR par amorces incluses avec pairs d’amorces universelles P1/P7 et R16F2n/R2. Les phytoplasmes ont été identifiés en tant que souches parentes de ‘Candidatus phytoplasma asteris’ avec un taux d’identité des nucléotides de plus de 99% par rapport à la souche de référence M30790, et ce, basé sur l’analyse in silico des fragments F2nR2 effectuée avec iPhyClassifier, l’outil en ligne. En se basant sur les recherches de l’homologie de séquence, le RFLP et les analyses phylogénétiques du gène 16S de l’ADNr, il a été possible de confirmer que les phytoplasmes, appartenant à deux lignées distinctes, aux sous-groupes 16SrI-A et 16SrI-B, infectaient le canola au Dakota du Nord. Une caractérisation supplémentaire des phytoplasmes, basée sur l’opéron de la protéine ribosomique et les gènes secY, a également confirmé la présence de deux lignées distinctes. La comparaison des séquences et les analyses RFLP basées sur les trois régions ont confirmé l’infection mixte causée par les phytoplasmes appartenant aux sous-groupes 16SrI-A et 16Sr1-B dans deux échantillons, un de 2011 et l’autre de 2012. À notre connaissance, il s’agit de la première mention du phytoplasme appartenant au sous-groupe 16SrI-A et d’une infection mixte causée par les phytoplasmes 16SrI-A et 16SrI-B chez le canola aux États-Unis.
Introduction
Aster yellows disease of canola and other Brassica species is associated with ‘Candidatus Phytoplasma asteris’ (Wang & Hiruki Citation2001; Olivier et al. Citation2006, Citation2010). Because of their obligatory parasitic nature, phytoplasmal aetiology is established using molecular methods based on several conserved regions; these include 16S rDNA, ribosomal protein (rp) operon (Lee et al. Citation2004) and protein translocase (secY) genes (Lee et al. Citation2006). Phytoplasmas are classified into groups and subgroups based on RFLP patterns resulting from sequence variations within these conserved regions. Among these, classification based on 16S rDNA is commonly used for identification purposes (Lee et al. Citation1998). ‘Candidatus Phytoplasma asteris’ is the reference strain for phytoplasmas in group 16SrI, which is considered to be the most diverse and widespread group and includes strains associated with more than 100 economically important diseases globally (Lee et al. Citation2004).
Canola (Brassica napus L.) plants showing typical Aster yellows (AY) symptoms, such as stunting, phyllody, leaf purpling, and formation of bladder-like siliques (Bertaccini et al. Citation1998), have been observed for several years in different regions of North Dakota. North Dakota is the largest producer of canola in the USA, accounting for ~80% of the crop production acreage (USDA-NASS Citation2014). One of the earliest reports of AY-symptomatic canola plants in North Dakota was made by Lamey et al. (Citation2000), who reported its occurrence in several fields but without quantifying it. Since then, AY-symptomatic canola plants have been observed sporadically at varying frequencies. The localized nature of the outbreaks may be misleading, however. For example, the state average incidence of AY-symptomatic plants in 2000 was 4.5%; however, in that year, eight out of the 16 counties represented some fields with incidences >5% (Lamey et al. Citation2001). Similarly, in 2007, the state-wide average incidence was 5.6% but approximately 22% of the 196 fields scouted had incidences >10% (Knodel, unpublished data). In 2012, a few canola fields were heavily affected by the disease in McLean and Cavalier counties, with incidences ranging between 10 and 20% (R. Beneda, personal comm.). Despite the continued presence of AY-symptomatic canola plants in North Dakota fields, the causal agent(s) associated with this disease in the state has not been confirmed. The objective of this study was to identify and characterize the pathogen(s) associated with AY in canola plants in North Dakota.
Plant material and nucleic acid extraction
Twenty canola plants showing typical AY symptoms were collected from three commercial fields in McLean County in 2011, and eight symptomatic plants originated from two experimental plots from the Research and Extension Center, North Dakota State University, in Langdon, Cavalier County in 2012. Approximately 100 mg of freeze-dried symptomatic field-derived and healthy greenhouse-grown canola tissues (leaves and inflorescences) were homogenized separately in 500 μL of Edwards buffer (Edwards et al. Citation1991) and total DNA was extracted following the phenol:chloroform:isoamyl alcohol procedure described by Ausubel et al. (Citation1995).
PCR analysis
Universal primer pairs P1/P7 (Schneider et al. Citation1995) and R16F2n/R16R2 (Gundersen & Lee Citation1996) specific for phytoplasmas were used for amplification of the 16S rDNA region in direct and nested PCR steps, respectively. Primer pairs rpF1/rpR1 (Lim & Sears Citation1992) and AYsecYF1/AYsecR1 (Lee et al. Citation2006) were used for PCR amplification of the ribosomal protein (rp) operon containing rpl22 and rps3 genes (producing a band ~1.2 kb in size), and the protein translocase gene (secY) (producing a band ~1.4 kb in size), respectively. All PCR reactions were performed in 20 μL reaction volumes containing 200 μM dNTPs each, 0.5 μM each of respective primers, and 1 U TopTaq DNA polymerase (Qiagen) and 1 μL of undiluted DNA. PCR was performed in an automated thermal cycler (PTC 100, MJ Research) with the following cycling conditions: initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing for 30 s (at 55°C for 16S rDNA and secY, 50°C for rp operon) and extension at 72°C for 1.5 min, and a final extension for 5 min at 72°C.
Sequence and RFLP analyses
The PCR products were purified using Wizard SV gel and PCR purification system (Promega, WI) and the DNA concentration was adjusted to 50 ng μL−1 before being sequenced on an ABI 3730 XL automated sequencer at Molecular Cloning Laboratories (MCLab, San Francisco, CA). Nucleotide homology search was conducted using the NCBI-BLAST (Altschul et al. Citation1990) to putatively identify phytoplasma strains. iPhyclassifier (Zhao et al. Citation2009) was used to further confirm the group and subgroup assignments based on BLAST search results of 16S rDNA sequences. To further confirm iPhyclassifier’s in silico RFLP analysis, 100 ng of the purified products of the nested PCR assay were digested separately with AluI, BfaI, BstUI, EcoRI, HaeIII, HhaI, HpaII, MseI, RsaI, Sau3AI and TaqI following the manufacturer’s instructions (New England Biolabs). RFLP analyses of rp and secY amplicons (100 ng) were performed by digesting with MseI, Tsp5091 and AluI. RFLP fragment patterns were visualized either in 3% or 5% super-fine resolution agarose (Agarose SFR™, Amresco Inc., OH) and compared with that of reference strains.
Phylogenetic analysis
Phylogenetic trees were constructed by parsimony based on the 16S rDNA sequences, as well as on the rp operon and secY gene sequences, using PAUP version 4.0 (Swofford Citation2002) as described in Lee et al. (Citation2006). Representative AY phytoplasma strains and other 16Sr groups were included for 16S rDNA (1338 bp) (Olivier et al. Citation2010), rp operon (1146 bp) (Lee et al. Citation2004) and secY (1326 bp) (Lee et al. Citation2006) as described in the original papers.
Results and discussion
The presence of 16S rDNA amplicons of ~1.8 kb in direct PCR assay using universal primer pairs P1/P7 and/or of ~1.2 kb in nested PCR assay using primers R16F2n/R16R2 specific for phytoplasmas confirmed the presence of a phytoplasma in the collected AY symptomatic canola samples. However, no amplicons were detected from healthy samples. BLAST results based on 16S rDNA sequences indicated that nine symptomatic samples collected in 2011 had 100% identity at the sequenced region and >99% nucleotide identity with that of GenBank accessions HM137306 and NR_074759 which belong to subgroup 16SrI-A (). In addition, 17 symptomatic samples, 10 collected in 2011 and seven collected in 2012, had >99% sequence homology with accessions AY265207 and AY265208 which belong to subgroup 16SrI-B (). However, sequences from two symptomatic samples, Bn11 collected in 2011 and Bn24 collected in 2012, were equally identical to GenBank accessions belonging to both subgroups, and thus could not be unambiguously assigned to either of them (). These two sequences had ambiguities in nucleotide sequences at key restriction sites that are used to distinguish 16Sr subgroups A and B. Based on their >99% nucleotide identity to the reference strain M30790 in the iPhyClassifier phytoplasma classification database, all 28 phytoplasmas from this study were identified as ‘Candidatus Phytoplasma asteris’ related strains. In congruence with BLAST results, an in silico RFLP analysis also classified nine samples as belonging to16SrI-A, 17 as 16SrI-B, and two samples as variants of 16SrI-B.
Table 1. Classification of 28 phytoplasma strains based on 16S rDNA, ribosomal protein and secY sequences into RFLP subgroups.
RFLP patterns of 16S rDNA sequences of the samples differed from each other only in restriction patterns created by digestion with BfaI, HhaI and MseI (). These results were in complete agreement with both BLAST and iPhyclassifier analyses, with nine and 17 samples identified as belonging to 16Sr subgroups IA and IB, respectively. Restriction patterns of samples Bn11 and Bn24 were identical to a pattern that would result from superimposing the 16SrI-A and 16SrI-B patterns (). Representative sequences from North Dakota samples belonging to 16Sr subgroups IA and IB have been deposited in GenBank and identified as strains NDBN-A (accession KF511801) and NDBN-B (accession KF511800), respectively. The presence of two 16SrI subgroups affecting canola separately or in mixed infections has been previously reported in Canada (Wang & Hiruki Citation2001; Olivier et al. Citation2010).
Fig. 1 RFLP analysis of 16S rDNA, rp operon and secY sequences of phytoplasma strains. a, RFLP profiles of the 16S rDNA nested PCR products amplified with primer pairs R16F2n/R2 and digested using restriction endonucleases BfaI, HhaI and MseI. b, RFLP profiles of the rp operon sequences (containing rpl22 and rps3 genes) amplified with primer pairs rPF1/rpR1 digested using restriction endonucleases AluI, Tsp5091 and MseI. c, RFLP profiles of the secY gene sequences amplified with the primer pairs AYsecYF1/AYSecYR1 using restriction endonucleases AluI, Tsp5091 and MseI. Lanes M is GeneRuler 1 kb plus DNA ladder (Life Technologies) molecular weight marker. Digests were separated on 3% super fine resolution agarose gel (SFR) except for MseI and Tsp5091 digests of rp and secY genes which were separated on 5% Agarose SFR gel. Samples Bn03 and Bn06: subgroup IA; Bn10 and Bn16: subgroup IB; Bn11 and Bn24: mixed infection.
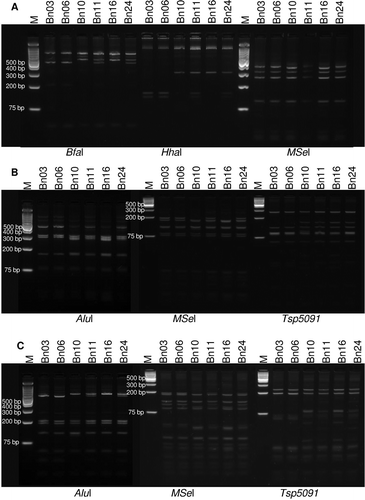
The observed ambiguity in nucleotide sequences and RFLP patterns of phytoplasma strains Bn11 and Bn24 could be the result of either a mixed infection or inter-operon heterogeneity between the rrnA and rrnB rRNA operons. RFLP and sequence analyses of two less conserved sequences, rp and secY, were used to determine the cause of this ambiguity. Sequence comparison indicated the presence of two distinct subgroups. The rp and secY sequences of strains NDBN-A and NDBN-B were deposited in GenBank with accession numbers KP796186-KP796187 and KP796188-KP796189, respectively. Similar to the 16S rDNA sequences, rp and secY gene sequences of Bn11 and Bn24 showed ambiguities at restriction enzyme recognition sites that distinguish subgroups A and B. This result was confirmed by the restriction profiles of the purified rp and secY amplicons (, ). The 16SrI-A and 16SrI-B phytoplasmas were identified as belonging to I-A and I-B subgroups, respectively, within their corresponding rp (Lee et al. Citation2004) and secY (Lee et al. Citation2006) RFLP subgroups. Similar to 16S rDNA profiles, the restriction patterns of rp and secY amplicons of Bn11 and Bn24 resembled the result of superimposition of restriction patterns of the two subgroups A and B, confirming the mixed nature of the infection.
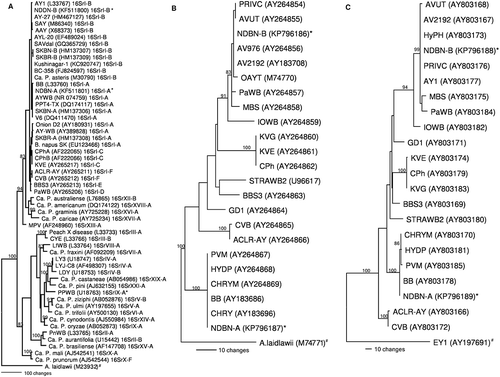
Based on phylogenetic analysis of 16S rDNA sequences, strains NDBN-A and NDBN-B were clustered within the phylogenetic lineage containing 16SrI-A and 16SrI-B strains, respectively (). Similarly, the analysis based on rp operon and secY genes clustered NDBN-A and NDBN-B phytoplasmas within the phylogenetic lineage containing only A (rpI-A, secY-IA) and B (rpI-B, secY-IB) subgroup strains, respectively (, ). Sequence comparisons, RFLP and phylogenetic analysis based on three regions therefore clearly indicated the presence of two distinct phytoplasma lineages infecting canola in North Dakota, in addition to a mixed infection by the two subgroups in two field samples. Occurrence of 16SrI-B phytoplasmas infecting canola in North Dakota were reported recently based on preliminary studies (Chittem & del Río Citation2013). To our knowledge, this is the first confirmed report of a 16SrI-A subgroup phytoplasma, and of a mixed infection by IA/IB subgroup phytoplasmas, in canola in the USA.
Fig. 2 Phylogenetic trees constructed by parsimony analyses based on three genetic regions. a, 16S rDNA (using representative sequences of several groups of phytoplasmas) b, rp operon (using representative sequences of the rp operon from AY subgroups) and c, secY (using representative sequences of the secY gene from AY subgroups) sequences using PAUP 4.0 version (Swofford Citation2002). Reference strains for 16S rDNA (Olivier et al. Citation2010), rp operon (Lee et al. Citation2004), and secY (Lee et al. Citation2006) were as described in the original papers. Accessions with an * represent the strains sequenced in this study and strains with # are used as outgroups. Trees were constructed via step-wise addition by 100 replicates of a heuristic search employing tree bisection and reconnection algorithm to find optimal trees. Branch lengths are proportional to the number of inferred character state transformations. Bootstrap analyses (1000 replicates) were conducted to determine the stability and support for the inferred clades. Bootstrap values above 80% are shown on main branches.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Ausubel F, Brent R, Kinston R, Moore D, Seidman JG, Smith J, Struhl K. 1995. Short protocols in molecular biology. 3rd ed. Unit 2.1: p. 2–3. New York, NY: Wiley and Sons.
- Bertaccini A, Vorácková Z, Vibio M, Fránová J, Navrátil M, Špak J, Nebesárová J. 1998. Comparison of phytoplasmas infecting winter oilseed rape in the Czech Republic with Italian Brassica phytoplasmas and their relationship to the aster yellows group. Plant Pathol. 47:317–324.
- Chittem K, del Río LE. 2013. Detection of ‘Candidatus Phytoplasma asteris’ in canola in North Dakota. Phytopathology. 103:S2.27 (abstr.).
- Edwards K, Johnstone C, Thompson C. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucl Acids Res. 19:1349.
- Gundersen DE, Lee IM. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopath Medit. 35:144–151.
- Lamey A, Knodel J, McKay K, Endres G, Fore Z, Andol K, Draper M. 2001. 2000 Canola disease survey in Minnesota, North Dakota and South Dakota. North Dakota State University Extension Service. Ext Rep. 63 p. 7.
- Lamey A, McKay K, Knodel J, Endres G, Andol K, Fore Z. 2000. 1999 Minnesota and North Dakota canola disease survey. North Dakota State University Extension Service. Ext Rep. 60 p. 5.
- Lee I-M, Gundersen-Rindal DE, Davis RE, Bartoszyk IM. 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. Int J Syst Bacteriol. 48:1153–1169.
- Lee I-M, Gundersen-Rindal DE, Davis RE, Bottner KD, Marcone C, Seemüller E. 2004. “Candidatus Phytoplasma asteris”, a novel phytoplasma taxon associated with aster yellows and related diseases. Int J Syst Evol Microbiol. 54:1037–1048.
- Lee I-M, Zhao Y, Bottner KD. 2006. SecY gene sequence analysis for finer differentiation of diverse strain in the aster yellows phytoplasma group. Mol Cell Probes. 20:87–91.
- Lim PO, Sears BB. 1992. Evolutionary relationships of a plant-pathogenic mycoplasmalike organism and Acholeplasma laidlawii deduced from two ribosomal protein gene sequences. J Bacteriol. 174:2606–2611.
- Olivier CY, Galka B, Séguin-Swartz G. 2010. Detection of aster yellows phytoplasma DNA in seed and seedlings of canola (Brassica napus and B. rapa) and AY strain identification. Can J Plant Pathol. 32:298–305.
- Olivier CY, Séguin-Swartz G, Hegedus D, Barasubiye T 2006. First report of ‘Candidatus Phytoplasma asteris’-related strains in Brassica rapa in Saskatchewan, Canada. Plant Dis. 90:832.
- Schneider B, Seemüller S, Smart CD, Kirkpatrick B. 1995. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas. In: Razin R, Tully JG, editors. Molecular and diagnostic procedures in mycoplasmology. Vol. 1. San Diego, CA: Academic Press; p. 369–380.
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, MA: Sinauer Associates.
- USDA-NASS. 2014. North Dakota Agriculture. Ag. Statistics No 83. August 2014. National Agricultural Statistics Service North Dakota Field Office, ND, USA.
- Wang K, Hiruki C. 2001. Molecular characterization and classification of phytoplasmas associated with canola yellows and a new phytoplasma strain associated with dandelions. Plant Dis. 85:76–79.
- Zhao Y, Wei W, Lee I-M, Shao J, Suo X, Davis RE. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int J Sys Evol Microbiol. 59:2582–2593.
