Abstract
In 2013, a new foliar disease was observed in processing lima bean (Phaseolus lunatus L.) plants in fields across western New York State, USA. The fungus Boeremia exigua var. exigua (Desm.) Aveskamp, Gruyter & Verkley was consistently isolated from lesions. The fungus was identified by characterization of morphological features and multiple genes (internal transcribed spacer, partial actin, β-tubulin, translation elongation factor 1-α and calmodulin) informative for speciation within the Didymellaceae. Pathogenicity was confirmed by inoculation of three lima bean cultivars in a repeated greenhouse experiment. Snap bean (Phaseolus vulgaris L.) and soybean (Glycine max (L.) Merr.), which are commonly encountered in the cropping rotation in New York State, were also inoculated, and demonstrated susceptibility to the pathogen. To the best of our knowledge, this is the first report of Boeremia exigua var. exigua causing tan spot disease on foliage of lima bean in New York State, USA.
Résumé
En 2013, une nouvelle maladie foliaire a été observée chez les plants de haricots de Lima de transformation (Phaseolus luna L.) dans les champs de l’ouest de l’État de New York, aux États-Unis. Le champignon Boeremia exigua var. exigua (Desm.) Aveskamp, Gruyter & Verkley a été invariablement isolé à partir de lésions. Le champignon a été identifié par caractérisation des traits morphologiques et de plusieurs gènes (espaceur transcrit interne, fragments d’actine, ß-tubuline, facteur d’élongation de la traduction 1-α et calmoduline), ce qui s’est avéré informatif sur le plan de la spéciation au sein de la famille des Didymellaceae. La pathogénicité a été confirmée par inoculation de trois cultivars de haricot de Lima à la suite d’expériences répétées en serre. Des haricots mange-tout (Phaseolus vulgaris L.) et des fèves de soya (Glycine max [L.] Merr.), qui sont couramment utilisés dans la rotation des cultures dans l’État de New York, ont également été inoculés, ce qui a permis de démontrer la vulnérabilité à l’agent pathogène. Pour autant que l’on sache, il s’agit de la première mention de l’apparition, dans l’État de New York, de la tache helminthosporienne sur le feuillage du haricot de Lima, causée par Boeremia exigua var. exigua.
Introduction
Approximately 650 hectares of lima beans (Phaseolus lunatus L.) are grown for processing in western New York State, USA. In 2013, a new disease affecting the foliage was observed in fields across the region. Symptoms of the disease were tan coloured, circular necrotic lesions with red-brown to brown coloured irregular margins (2–19 mm in diameter) on leaves. Lesions were usually discrete, but occasionally coalesced to form larger blotches (). Dark coloured pycnidia could be observed in the necrotic centres of older lesions. The primary objective of this study was to evaluate the role and importance of a pycnidial fungus consistently associated with the lesions, to assess its pathogenicity to three common cultivars of lima bean, and to determine its role in the foliar disease observed in commercial fields. Confirmation of pathogenicity and evaluation of the ability of the fungus to cause disease on snap bean and soybean, common legume rotational crops, was also established.
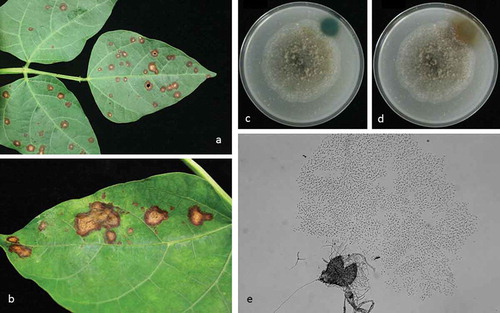
Fig. 1 (Colour online) Disease symptoms caused by Boeremia exigua var. exigua on lima bean and fungal morphology. a, Discrete tan spot necrotic lesions on lima bean field sample. b, coalescence of necrotic tan spot lesions on a lima bean field sample. c, reaction of Boeremia exigua var. exigua on oatmeal agar to 5 M NaOH at 20 min after application; and d, reaction to 5 M NaOH at 120 min after application; (e) pycnidium and conidia at ×400 magnification.
Materials and methods
Sampling and pathogen isolation
Diseased leaves were sampled in 2013 from three commercial lima bean fields in western New York State, USA. Margins of the lesions were excised, surface sterilized in 10% household bleach for 30 s, and rinsed three times in sterile distilled water. The excised tissue (<2 mm3) from the junctions of healthy and necrotic areas were placed onto 2% water agar (WA) in Petri plates and incubated at room temperature under 12 h fluorescent lighting. Upon growth of fungi from the plated tissue, a small plug containing 3–4 hyphal tips was excised, placed on a fresh 2% WA plate and incubated for 24 h at room temperature. After incubation, a single growing mycelial tip was excised using a sterile scalpel blade and placed onto potato dextrose agar (PDA). Transfers were later made to malt extract (MEA) and oatmeal agars (OA). All isolates underwent hyphal tipping prior to use in further studies of morphological and molecular characteristics. Pycnidial and conidial morphology of isolates were observed after 30 days. The shape, length and width of 50 conidia were measured at 400× magnification. The presence of metabolite ‘E’ was tested on OA after 7 days by observing the reaction of 5 M NaOH (Boerema et al. Citation2004).
Molecular characterization
Three individual hyphal-tipped isolates obtained from diseased lima bean leaves putatively identified as B. exigua var. exigua on the basis of morphology were selected for molecular characterization and grown in potato dextrose broth for 7 days. Mycelia (~100 mg) were collected using vacuum-filtration and ground in a TissueLyserII system (Qiagen Inc., Valencia, CA) for 45 s at 60 hz s−1 using two 4.5 mm tungsten carbide beads. Genomic DNA was extracted with a DNeasy Plant Mini Kit (Qiagen Inc.). After extraction, DNA was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). DNA was diluted to approximately 25 ng µL−1 and stored at −20°C. The internal transcribed spacers (ITS) and 5.8S ribosomal DNA was amplified using primers ITS1 (5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ) and ITS4 (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) (White et al. Citation1990). Amplification conditions consisted of an initial denaturation at 94°C for 5 min, followed by 29 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min. A final elongation step of 72°C for 10 min was used. PCR reactions contained 1 µL of each 10 mM primer, 1 µL of 10 mM dNTP, 0.25 µL Taq polymerase, 5 µL New England Biolabs 10× Standard Taq buffer, and 6 µL of genomic DNA. The total volume of the reaction was adjusted to 50 µL using ddH2O.
The partial actin, β-tubulin, translation elongation factor 1-α (TEF) and calmodulin regions were also amplified using an identical reaction recipe. The partial actin gene was amplified with primer set ACT-512F (5ʹ-ATGTGCAAGGCCGGTTTCGC-3ʹ) and ACT-783R (5ʹ-TACGAGTCCTTCTGGCCCAT-3ʹ) (Carbone & Kohn Citation1999) with the PCR protocol described by Aveskamp et al. (Citation2009). For the TEF gene, primer set EF1-983F and EF1-1567R (http://www.aftol.org/pdfs/EF1primer.pdf) was used within the protocol described by Vaghefi et al. (Citation2012). For the β-tubulin gene, primer set BT2F (5ʹ-GTBCACCTYCARACCGGYCARTG-3ʹ) and BT4R (5ʹ-CCRGAYTGRCCRAARACRAAGTTGTC-3ʹ) (Woudenberg et al. Citation2009) with PCR protocol from Aveskamp et al. (Citation2009) was used. For the calmodulin gene, primer set CAL228F and CAL2Rd (Carbone & Kohn Citation1999; Groenewald et al. Citation2013) were used with an amplification protocol consisting of an initial denaturation at 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, followed by a final elongation step of 72°C for 5 min. Amplicons were sequenced at the Cornell University Biotechnology Resource Centre (Ithaca, NY, USA) in both directions and consensus sequences assembled.
Resultant sequences were compared with Didymellaceae reference isolates deposited within the GenBank database of the National Centre for Biotechnology Information. Sequences and those retrieved from GenBank were aligned using MUSCLE (Edgar Citation2004), and trimmed by hand to the same length and aligned. Phylogenetic analyses were performed with concatenated sequences of the five genes with reference isolates within the Didymellaceae. Bayesian analysis was conducted using MrBayes v3.2.2 (Ronquist & Huelsenbeck Citation2003), with the HKY85 model with gamma correction within the Geneious version 8 (http://www.geneious.com; Kearse et al. Citation2012) software program. The analysis consisted of four chains (one cold, three heated) and were run for 1 100 000 generations, with a burn-in length of 100 000 generations and a random seed. Trees were sampled every 200 generations. A majority rule consensus tree was generated and labelled with consensus support as a percentage. Phoma paspali CBS 560.81 was used as an outgroup for this analysis.
Pathogenicity of Boeremia exigua var. exigua to lima bean
A single isolate was selected from the three genetically identical isolates for use in pathogenicity testing. Inoculation of the isolate to detached lima bean leaves placed in Petri dishes provided preliminary evidence of pathogenicity (data not shown). A pathogenicity trial was conducted in the greenhouse and repeated once. Three-week-old lima bean plants at growth stage V2 (cultivars ‘Cypress’, ‘Maestro’ and ‘Kingston’) were inoculated with 5 mL of a conidial suspension (3 × 105 conidia mL−1 with 0.2% Tween-20) prepared from a 28-day-old culture, using a hand sprayer. Germination efficiency (%) was quantified by spreading 30 µL of inoculum across the surface of a PDA plate. Plates were then incubated at room temperature for 24 h. Fifty conidia were examined at 400× magnification to assess viability. A conidium was considered germinated if the germ tube was at least half of the length of the conidium.
Eight plants of each cultivar were inoculated with a B. exigua var. exigua isolate within each trial. In addition, eight plants of each cultivar received the same quantity of distilled water with 0.2% Tween-20 as non-inoculated controls, within each trial. Immediately after inoculation, plants were covered with clear plastic bags for 5 days at approximately 20°C in the greenhouse. Plants were examined at 25 days post-inoculation for disease incidence (number of plants showing symptoms/total number of inoculated plants × 100) and severity (number of diseased leaves/total number of leaves per plant × 100). Isolations (n = 18) were conducted from lesions that developed on inoculated plants using the technique as described for field samples. Data from both trials were combined to analyse the effect of inoculation and cultivar on disease incidence and severity using generalized linear modelling (R Studio, version 8; R Core Team Citation2014). Data were log-transformed prior to analysis. Means were separated using Fisher’s protected least significant difference test (P = 0.05).
The pathogenicity of a B. exigua var. exigua isolate from lima bean to other beans commonly encountered in the cropping rotation was quantified by inoculation to snap bean (Phaseolus vulgaris L.) ‘Huntington’, ‘Masai’ and ‘Caprice’, and soybean (Glycine max (L.) Merr.) ‘92Y51’ and ‘92Y38’ in an additional greenhouse trial. Methods for inoculation preparation, application, plant incubation and disease assessments were identical to those described for the lima bean pathogenicity test. Eight plants of each cultivar were inoculated with a conidial suspension as described above. In addition, seven plants of each cultivar received the same quantity of distilled water with Tween-20 as non-inoculated controls. Eight lima bean plants ‘Cypress’ were also included in the trial as positive controls. The pathogenicity trial to snap bean and soybean cultivars was performed once. Isolations (n = 15–20) were conducted from lesions that developed on inoculated plants using the technique described for field samples. Data were analysed to determine the effect of inoculation and species on disease incidence and severity using generalized linear modelling (R Studio version 8; 2014).
Results and discussion
Isolations and identification
Isolations resulted in a fungus that on OA produced colonies with irregular margins and appeared grey to dark grey with younger white aerial mycelia. Colonies on OA were 4.7–6.1 cm in diameter after 7 days and had a strong colour change reaction to 5 M NaOH, indicating the production of metabolite ‘E’ (, ). On MEA, colonies had highly irregular margins and appeared grey with younger white to cream aerial mycelia. Colonies were 4.8–6.3 cm in diameter after 7 days on MEA and conidia were hyaline, aseptate and oblong, ranging from 3.9–6.4 × 1.7–3.1 µm (). Morphological characteristics and the presence of metabolite ‘E’ suggested this fungus was Boeremia exigua var. exigua (Desm.) Aveskamp, Gruyter & Verkley, comb. nov. [basionym: Phoma exigua Desm., Ann. Sci. Nat. Bot. III 11:282 (1849)].
The sequences amplified from the ITS region, partial actin, β-tubulin, TEF and calmodulin genes were 98–100% identical to B. exigua var. exigua (GenBank accession numbers FJ427001, EU880846, FJ427112, JF9253281 and AY831533, respectively). Phylogenetic analysis using the five concatenated sequences showed that all three isolates were identical to each other and grouped with the B. exigua var. exigua type isolate (CBS 431.74; ). Our isolate has been deposited in the United States Department of Agriculture – Agricultural Research Service Culture Collection (NRRL accession number 66227). Sequences for the ITS, TEF, partial actin and β-tubulin have been deposited in GenBank (accession numbers KP746412, KP746413, KP746414 and KP746415, respectively).
Fig. 2 Phylogenetic relationships inferred using a Bayesian inference dendrogram between Boeremia exigua var. exigua identified in this study (presented in bold) and other Didymellaceae using concatenated sequences of the internal transcribed spacer region (ITS), partial actin (ACT), β-tubulin (TUB), translation elongation factor (TEF), and partial calmodulin (CAL) genes. CBS-KNAW Fungal Biodiversity Centre reference numbers are provided following species name. Numbers at branches indicate percentage bootstrap support. Scale bar indicates the proportional genetic similarity. The dendrogram is rooted using Phoma paspali CBS 560.81.

Pathogenicity testing
Conidial germination of inoculum used in all pathogenicity trials ranged from 83 to 94%. Necrotic lesions were visible on inoculated lima bean plants within 25 days, resulting in higher disease incidence and severity compared with the non-inoculated controls (P < 0.001; ). The mean disease incidence and severity in inoculated plants was 85.4% and 11.4%, respectively, across the three cultivars (). Lima bean cultivar had no significant effect on disease incidence (P = 0.99) or disease severity (P = 0.661; ). The reisolation frequency of B. exigua var. exigua was 83–100%. A small amount of disease was observed in non-inoculated plants, due to inadvertent spore spread in the greenhouse.
Table 1. Effect of inoculation with Boeremia exigua var. exigua on foliar disease severity (%) in lima, snap and soybean across two trials conducted in the greenhouse. Data were log transformed before analysis and means reported here are back-transformed.
Table 2. Effect of inoculation with Boeremia exigua var. exigua on disease incidence and severity (%) on lima bean, snap bean and soybean cultivars. Experiment 1 reports analysis of combined data from two greenhouse trials. Experiment 2 reports data from a single greenhouse trial. Data were log-transformed before analysis and are reported herein as back transformed.
Disease symptoms similar to those observed in the field and greenhouse-inoculated plants were obtained on snap and soybean plants following inoculation with B. exigua var. exigua. The reisolation frequency ranged from 80 to 93% for all cultivars tested. Species and cultivar had no significant effect on disease incidence (P = 0.56) but had a significant effect on disease severity (P = 0.001; ), suggesting that some soybean and snap bean cultivars (e.g. snap bean cultivar ‘Masai’) tested were more susceptible to B. exigua var. exigua.
To the best of our knowledge, this is the first report of B. exigua var. exigua causing foliar disease on lima bean in New York State, USA. The pathogen is likely to contribute to foliar diseases affecting lima bean productivity in this region and pathogen spread may have been exacerbated by wet and humid field conditions during the sampling year. Boeremia exigua var. exigua (syn. Phoma exigua var. exigua; Aveskamp et al. Citation2010) has been demonstrated to be the causal agent of Ascochyta leaf spot on snap bean (Bardas et al. Citation2008) and associated with foliar disease in soybean (Boerema et al. Citation2004; Kövics et al. Citation2014). The pathogenicity of B. exigua var. exigua isolates causing foliar disease on lima bean, snap bean and soybean may have implications for cropping rotation decisions for disease management. Further work will focus on quantifying the effect of this disease on lima bean productivity in New York State.
Acknowledgements
This work was supported by The New York State Agricultural Experiment Station Summer Scholars programme and a Cornell University Graduate Student Extension Assistantship for the first author and the New York State Vegetable Research Association and Council. Thanks to Carol Bowden, Cornell University for excellent technical assistance and Dr Frank Hay, Cornell University, for constructive comments on an earlier draft of the manuscript.
References
- Aveskamp MM, de Gruyter J, Woudenberg JHC, Verkley GJM, Crous PW. 2010. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol. 65:1–60.
- Aveskamp MM, Verkley GJM, de Gruyter J, Murace MA, Perelló A, Woudenberg JHC, Groenewald JZ, Crous PW. 2009. DNA phylogeny reveals polyphyly of Phoma section, Peyronellaea and multiple taxonomic novelties. Mycologia. 101:363–382.
- Bardas GA, Tziros GT, Tzavella-Klonari K. 2008. First report of Ascochyta leaf spot caused by Phoma exigua var. exigua on common bean in Greece. Plant Dis. 92:653.
- Boerema GH, de Gruyter J, Noordeloos ME, Hamers MEC. 2004. Phoma identification manual: differentiation of specific and infra-specific taxa in culture. Oxon (UK): Wellingford.
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 91:553–556.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 32:1792–1797.
- Groenewald JZ, Nakashima C, Nishikawa J, Shin H-D, Park J-H, Jama AN, Groenewald M, Braun U, Crous PW. 2013. Species concepts in Cercospora: spotting the weeds among the roses. Stud Mycol. 75:115–170.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kövics GJ, Sándor E, Rai MK, Irinyi L. 2014. Phoma-like fungi on soybeans. Crit Rev Microbiol. 40:49–62.
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Vaghefi N, Pethybridge SJ, Ford R, Nicolas ME, Crous PW, Taylor PWJ. 2012. Stagonosporopsis spp. associated with ray blight disease of Asteraceae. Aust Plant Pathol. 41:675–686.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Spinsky TJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press; p. 315–322.
- Woudenberg JHC, Aveskamp MM, de Gruyter J, Spiers AG, Crous PW. 2009. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 22:56–62.
