Abstract
Little is known about the molecular interaction of wheat and leaf rust (Puccinia triticina Eriks). However, genomic tools are now becoming available so that the host–pathogen interactions can be better understood. Significant efforts are being placed on understanding the secretomes of various pathogens as secreted peptides are believed to be the best candidates for avirulence effectors. In this work, a P. triticina haustorial cDNA library was evaluated for the presence of proteins containing secretion signals. Ten predicted proteins were found in the library, of which two were expressed in haustorial fractions. Three of the secreted proteins, Pt3, Pt12 and Pt27, were used in biolistic experiments to determine whether they could induce hypersensitive cell death, which is commonly observed in incompatible rust interactions with wheat leaf rust resistance genes. When Pt3 was co-bombarded with a β-glucoronidase (GUS)-expressing vector into wheat isolines with resistance genes Lr9 or Lr24, a significant reduction of GUS expression was observed, presumably due to hypersensitive cell death. In other co-bombardment experiments, Pt27 induced a significant reduction in GUS expression in the Lr26 isoline. These results suggest that Pt3 and Pt27 may function in avirulence against wheat leaf rust in resistant genotypes.
Résumé
Nous en savons peu sur l’interaction moléculaire blé-rouille des feuilles (Puccinia triticina Eriks). Toutefois, nous avons maintenant à notre disposition des outils génomiques qui nous permettent de mieux comprendre les interactions hôtes-agents pathogènes. Des efforts considérables sont consacrés à la compréhension des sécrétomes de divers agents pathogènes, dont les peptides sécrétés semblent être les meilleurs candidats en ce qui a trait aux effecteurs d’avirulence. Au cours de cette étude, une banque d’ADNc d’haustoriums de P. triticina a été examinée pour y détecter des protéines contenant des signaux de sécrétion. Nous avons trouvé dix protéines sécrétées dans la banque, dont deux étaient exprimées en fractions haustoriales. Trois des protéines sécrétées, Pt3, Pt12 et Pt27, ont été utilisées dans des expériences basées sur la biolistique afin de déterminer si elles pouvaient induire la mort cellulaire hypersensible qui est couramment observée lors d’interactions incompatibles de la rouille avec des gènes de la résistance à la rouille des feuilles chez le blé. Lorsque la protéine Pt3 était projetée avec un vecteur exprimant la β-glucoronidase (GUS) dans des lignées isogéniques possédant les gènes de résistance Lr9 ou Lr24, nous avons observé une réduction significative de l’expression de la GUS, sans doute causée par la mort cellulaire hypersensible. Au cours d’autres expériences de projection, la protéine Pt27 a induit une réduction significative de l’expression de la GUS dans la lignée isogénique Lr26. Ces résultats suggèrent que les protéines Pt3 et Pt27 peuvent engendrer une réaction d’avirulence quant à la rouille des feuilles chez les génotypes résistants.
Introduction
Puccinia triticina Eriks is an obligate biotrophic pathogen that is the causal agent of leaf rust in wheat (Triticum aestivum L.). Leaf rust is the most severe wheat disease in Kansas (Appel et al. Citation2009) and resistant varieties are used as an effective way of disease control. There are more than 70 wheat leaf rust (Lr) resistance genes that have been named and characterized (McIntosh et al. Citation2010). Most provide resistance in a gene-for-gene manner (Flor Citation1955), though some provide broad-spectrum resistance (McIntosh et al. Citation2010). Because of the monoculture of wheat, and the large acreages of a single cultivar, significant selection pressure is applied to the fungus to overcome resistance. Asexual urediniospores containing virulence changes are then cyclically produced, dispersed by wind, and can cause serious epidemics. New races are collected each year in the US Great Plains (Ordoñez & Kolmer Citation2009); however, the sexual host for P. triticina is absent and the source of genetic variation is not understood.
Among the biotrophs, the best-characterized pathosystem is flax (Linum usitatissimum L.) rust caused by Melampsora lini Ehrenb. Lev. Flor (Citation1955) which was the first to show that the resistance in flax and the inability of rust races to infect are genetically based in the plant and in the pathogen. Wheat leaf rust behaves in a similar manner. Avirulence (Avr) is either dominant or semi-dominant and is dependent on the respective host resistance gene (Dyck & Samborski Citation1968). This type of resistance is now known as effector triggered immunity (ETI; Jones & Dangl Citation2006). Pathogen effectors are secreted to suppress plant immunity, modify plant physiological responses to enable pathogen infection (Petre et al. Citation2014) and for inducing the uptake of nutrients (Catanzariti et al. Citation2010; Garnica et al. Citation2014). During ETI, certain pathogen effectors are perceived by plant disease resistance proteins, and through a signalling cascade, a host response is triggered to prevent disease. Changes in either the effector or the resistance protein may yield a compatible reaction. Numerous unique effectors from filamentous phytopathogenic fungi have been cloned. Most are small proteins that have a secretion signal at the amino terminal end of the protein (De Wit et al. Citation2009; Rafiqi et al. Citation2012).
Genetic and genomic approaches have been used to clone avirulence effectors. Pwl1, Pwl2 and AvrPita from Magnaporthe oryzae Couch and AVRL567 from M. lini (Dodds et al. Citation2004) were cloned using map based cloning (Valent et al. Citation1986; Sweigard et al. Citation1995; Orbach et al. Citation2000). Rust Transferred Protein 1 from Uromyces fabae (Pers.) J. Schröt. (UfRTP1) (Hahn & Mendgen Citation1997), AVRP123, AVRP4 and AVRM from M. lini (Catanzariti et al. Citation2006) were isolated by predicting secreted proteins from haustorial cDNA libraries. Similar approaches have been done in the wheat rusts. Yin and collaborators (Citation2009) generated haustorial ESTs from wheat stripe rust (Puccinia striiformis Westend) and 15 genes were predicted to encode secreted proteins. Fifty proteins with secretion signals were found in a proteome analysis of P. triticina haustoria (Song et al. Citation2011). Next-generation sequencing of RNA from infected tissue identified 456 cDNA sequences common in six P. triticina races with secretion signals (Bruce et al. Citation2014). However, none have been verified as avirulence factors.
Genomic resources are rapidly becoming available for cereal rusts with the release of the P. graminis, P. triticina and P. striiformis genomes (Duplessis et al. Citation2011; Zheng et al. Citation2013; http://www.broadinstitute.org/annotation/genome/puccinia_group, Broad Institute, Cambridge, MA). However, because they are obligate biotrophs, functional validation of avirulence genes is a challenging task. Transformation of the fungus by bombardment (Webb et al. Citation2006) or by Agrobacterium (Lawrence et al. Citation2010) can be used to evaluate candidate mutants and validate the avirulence, though selecting specifically for transformed spores can be difficult. Alternatively, transient expression experiments in host plants can be used to characterize effectors. Particle bombardment in isogenic lines is frequently used to co-express the candidate Avr effector proteins along with a reporter gene (Jia et al. Citation2000; Qutob et al. Citation2002; Allen et al. Citation2004; Armstrong et al. Citation2005; Rehmany et al. Citation2005; Ridout et al. Citation2006; Dou et al. Citation2008; Kaneda et al. Citation2009). After bombardment into resistant leaves, the transiently expressed Avr protein will induce cell death via recognition by the cognate R gene. A reduction of reporter gene expression, such as green fluorescence protein (GFP) or ß-glucuronidase (GUS), is used to quantify the resistance response as an indication of cell death in an AVR-R recognition manner (Kale et al. Citation2010).
The wheat-leaf rust pathosystem is poorly characterized at the molecular level. There are wheat isogenic lines that facilitate the study of pathogenicity of the fungus toward specific Lr genes (McIntosh et al. Citation1995) and enable characterization for Avr function in given rust isolates. To date, only two candidate avirulence genes have been characterized in the cereal rusts (Nirmala et al. Citation2011; Upadhyaya et al. Citation2014); however, formal confirmation of these genes as actual avirulence effectors has not been reported. The identification of leaf rust Avr genes may provide information to generate new strategies for disease control. In this work, we have further characterized a haustorial cDNA library previously included in a transcriptome survey of P. triticina (Xu et al. Citation2011). Ten proteins with secretion signals were characterized for expression during infection and three were tested for recognition by host resistance genes. We report from transient expression experiments that two of the three proteins induced a reduction of ß-glucoronidase expression in wheat lines containing resistance genes, suggesting they have a role in avirulence.
Materials and methods
Plant material and rust culture
Seedlings of the susceptible wheat cultivar ‘Wichita’ were grown in pans (7.5 cm2) containing Metro Mix 360 soil medium (Sun Gro, Bellevue, WA) in a growth chamber with a 16 h photoperiod, 21ºC, and light levels of 145 mol m−2 s−2. At the 2–3 leaf stage, plants were inoculated with 30 mg of urediniospores of P. triticina race PBJL (avirulent on Lr2a, Lr3 ka, Lr9, Lr10, Lr16, Lr14a, Lr18, Lr24, Lr26, Lr30; virulent on Lr1, Lr2c, Lr3a, Lr11, Lr17, LrB) suspended in 2 mL of Soltrol 170 isoparaffin solvent (Chevron Phillips Chemical Co, The Woodlands, TX). Inoculated plants were incubated overnight in a 100% humidity chamber at 18ºC. Plants were transferred back to the growth chamber at the conditions listed above.
Haustoria isolation, cDNA cloning and sequencing
Haustoria were isolated by following the protocol developed by Hahn & Mendgen (Citation1992). Heavily infected leaf tissue was harvested at 6 days post inoculation (dpi) and washed in deionized water. Eight grams of infected leaves were placed in 100 mL of ice-cold homogenization buffer (0.3 M sorbitol; 20 mM MOPS, pH 7.2; 0.1% bovine serum albumin (BSA); 0.2% 2-mercaptoethanol and 0.2% PEG 6000) and homogenized in a Waring blender at maximum speed for 15 s. The suspension was filtered through a 20-µm nylon mesh, rinsed with homogenization buffer, divided into four 50-mL centrifuge tubes, and centrifuged in a JA-18 rotor at 5000–7000 × g for 10 min. The pellet was re-suspended in 8 mL of ice-cold suspension buffer (0.3 M sorbitol; 10 mM MOPS, pH 7.2; 0.2% BSA; 1 mM KCl; 1 mM MgCl2). The preparation was centrifuged at 5000–7000 × g for 10 min and the pellet re-suspended completely in 4 mL suspension buffer. Two column volumes of re-suspended material was loaded onto a column with CNBr-activated sepharose 6MB beads (Sigma Aldrich, St. Louis, MO) and allowed to sit for 15 min. The column was washed by overlaying 2 column volumes of suspension buffer and rinsed five times. The column outlet was closed and one void volume of suspension buffer was added and the column content was agitated by pipetting. The sepharose beads were allowed to settle for 1–2 min and the haustoria containing supernatant was transferred to a 1.5 mL tube. Haustoria were pelleted at 15 000 × g for 1 min in a microfuge. cDNA library preparation, sequencing and contig assembly were previously described in Xu et al. (Citation2011).
Sequence analysis and database searches
The 188 assembled haustorial ESTs (Xu et al. Citation2011) were aligned to the P. triticina genome database (http://www.broadinstitute.org/annotation/genome/puccinia_group/Blast.html), using the settings for BLAST alignment of BLOSUM62, FILTER (YES), alignment type = gapped, and threshold of e −3 (Altschul et al. Citation1997). BLAST2GO (Conesa et al. Citation2005) was used for functional annotation of the ESTs with the settings of QBLAST-NCBI low complexity filter, annotation cut-off of 55, and GO weight of 5. Sequences were screened for repetitive elements using CENSOR and default settings (Kohany et al. Citation2006). Open reading frames (ORF) were identified with FGENESH (Softberry, Mount Kisco, NY). Identified ORFs were analysed for the presence of a predicted nuclear localization signal using PredictNLS (Cokol et al. Citation2000) with default settings, and scanned for motifs using MEME Suite of motif-based sequence analysis tools (Bailey et al. Citation2009). Settings for the searches were: optimum number of sites (more than 2 and less than 100), occurrence of a single motif distributed among the sequences as (any number of sequences), and maximum of 6 motifs per sequence. Secondary structure comparison was done with LOOPP@BioHPC (http://cbsuapps.tc.cornell.edu/loopp.aspx) with default settings. Predicted secreted proteins were identified with the program SignalP v. 3.0 (Bendtsen et al. Citation2004) with the settings: Organism group: Eukaryotes; output format: standard; Methods: Neural Network (NN) and Hidden Markov models (HMM); graphics: GIF (inline); output format: standard. Score values from HMM and NN ≥ 0.6, were selected as positive for secretion signal.
PCR amplification and cloning of candidate genes
Primers were designed to amplify the full-length coding sequence of each putative secreted peptide and included a BamHI restriction site (Supplementary ). Forward primers included the start ATG codon and the following 19 bases. Reverse primers included the last 19 bases of the coding region and the termination codon. Escherichia coli bacteria containing pGEM T Easy vectors with the full-length coding sequence of the Avr candidates, as determined by alignment to the genome, were pulled from the haustoria cDNA library and cultured overnight on LB plates containing 100 mg L−1 ampicillin at 37°C. Genes that did not have full length clones available were cloned from cDNA of infected tissue. Two mL cultures of single colonies were then grown overnight and plasmid DNA preparations were made using Qiagen miniprep kit (Qiagen, Valencia, CA). PCR conditions were 20 µL reactions containing 2 µL of plasmid DNA (250 ng), 10 pmol of both forward and reverse primers for each candidate, 2.0 µL 10× Taq reaction buffer (Sigma), 2.5 mM MgCl2, 2.5 mM dNTPs, and 1 µL of Taq enzyme (Sigma). Amplification conditions on the MJ Research PTC100 consisted of 92°C for 3 min, then 35 cycles of 92°C for 1 min, 60°C for 2 min, and 72°C for 2 min, and one cycle of 72°C for 10 min. The amplicons were ligated into the TA vector, pCR2.1, using TA cloning kit (Invitrogen, Carlsbad, CA) following manufacturer's instructions.
Table 1. Predicted secreted proteins identified in P. triticina haustoria-specific expressed sequence tags (ESTs). EST sizes are in base pairs (bp) and translated amino acids (aa). Proteins were characterized for number of cysteine residues (Cys), secretion signal peptide (SP) length and Signal P score.
The plant expression vector pAHC17 (Christensen & Quail Citation1996) was used for transient expression of the candidate Avr factors. The genes were initially amplified by PCR as described above, ligated into the pCR2.1 TA cloning vector (Invitrogen) and transformed into INValphaF E. coli cells. Inserts were cleaved from the plasmid using BamHI, gel-purified using the Gel Extraction Kit (Qiagen) and cloned into the BamHI site of pACH17. All candidate Avr expression vectors were sequenced to verify sequence and orientation.
Semi-quantitative RT-PCR
To evaluate the expression of selected candidate genes during infection, total RNA was isolated from uninfected seedling wheat leaves at the 2–3 leaf stage, infected wheat leaves harvested 1, 2, 3, 4, 5 and 6 dpi; in vitro germinated urediniospores (Webb et al. Citation2006) at 30 min, 1, 2 and 3 h, and isolated haustoria. cDNA was made using First-Strand cDNA Synthesis SuperScript II RT (Invitrogen) following the manufacturer’s protocol. Primers (Supplemental ) for gene amplification were as above. Expression control primers were designed for the P. triticina ß-tubulin (PTTG_00759.1) to validate cDNA quality and PCR reaction success. The amplicons were visualized on a 1% agarose gel in 1×TAE buffer.
Transient expression in isogenic lr lines
Puccinia triticina race PBJL induces a strong hypersensitive response in ‘Thatcher’ isogenic lines Lr9, Lr24, Lr26 and Lr52. Putative Avr candidates were co-bombarded with the reporter gene, ß-glucuronidase (UidA) construct into ‘Thatcher’ isogenic lines carrying resistant genes Lr 9 (RL6010), Lr 24 (RL6064), Lr 26 (RL6078) and Lr52 (RL6107). ‘Thatcher’ (Tc) was used as a control. In vitro grown plants were used as a source of tissue. Seeds were sterilized with 96% ethanol for 1 min and washed once with sterile ddH2O for 1 min. The seed was then treated with a 20% hypochlorite solution for 20 min, followed by three washes with sterile ddH2O of 1 min each. Seeds were dried overnight at room temperature in a laminar flow hood and stored at 4ºC. Pre-germination of the seeds was necessary to coordinate the time of germination. Twenty seeds were placed into a Petri dish containing Whatman 1 paper (90 mm) saturated with sterile ddH2O. They were kept at 4ºC for 48 h and subsequently transferred to room temperature for 2 days. Individual seeds were placed on the surface of 0.5 × Murashige and Skoog (Citation1962) agar solution (Murashige & Skoog salt, 2.15 g L−1 (Sigma-Aldrich); sucrose 15 g L−1; phytagel 1 g L−1; pH 5.7) contained in a 25 ×150-mm glass test tube. Tubes were placed in a growth chamber at 21ºC and 16 h photoperiod and photon flux density of 145 mol m−2 s−1. After 2 weeks, the first expanded leaves were cut into 10 cm long explants and cultured for 48 h in a Petri dish containing 0.5× MS medium, with conditions previously described.
DNA gold carrier particles were prepared by taking 30 µL of suspended DNAdelTM Gold Carrier Particles (S550d; Seashell Technology, La Jolla, CA) and mixing them with 20 µL of binding buffer provided by the kit, for a final concentration of 30 mg mL−1 of gold particles. 2.5 µL of GUS plasmid pAHC27 (1 µg µL−1) and 2.5 µL of candidate plasmid (1 µg µL−1) were added to the gold suspension for a total volume of 55 µL. Equal volume of precipitation buffer was added and incubated a room temperature for 3 min, followed by a centrifugation at 13 000 × g for 10 s. The pellet was washed with 500 µL of 100% ethanol and vortexed. The suspension was centrifuged again, the ethanol discarded, and the pellet was resuspended by adding 75 µL of 100% ethanol. The solution was sonicated for 10 min to break up the gold clumps (Barnstead Lab-Line, Aqua wave 9377, St. Louis, MO) and 7.5 µL of the suspension was placed onto a macrocarrier disk (Bio-Rad, Inc., Hercules, CA) and allowed to air-dry.
Two pre-cultured leaves 10 cm long were placed in a Petri dish containing wet filter paper and they were held in place by an aluminium disc (Supplementary ). The Bio-Rad PDS-1000/He particle gun device (Bio-Rad), modified with a focusing barrel attachment (Torisky et al. Citation1996) was used for bombardment. The Petri dish containing the two leaves was placed on the stage at 9 cm. The chamber vacuum was 25 in Hg and rupture disks of 1100 psi (Bio-Rad) were selected. After bombardment, leaves were again cultured in Petri dishes containing 0.5× MS medium for 48 h in a growth chamber as described above. The experimental design was two leaves per bombardment, 10 for the resistant line and 10 for the ‘Thatcher’ control, per avirulence candidate. The experiment was repeated twice.
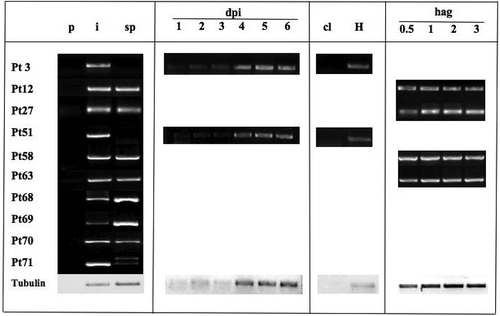
Fig. 1 Validation of predicted secreted proteins from P. triticina selected as effector candidates using RT-PCR. RNA isolated from non-infected plants (p), heavily infected plant tissue 6 days post inoculation (i) and germinated spores at 16 h (sp). Time course expression was evaluated from 1 to 6 days post-inoculation (dpi) and from spores 0.5 to 3 h after germination (hag). Total RNA from haustoria preparation (H) and mock inoculated plants (cl).
Bombarded leaves were cut in 2 cm long pieces that included the bombarded area and placed in GUS buffer (100 mM Na phosphate buffer, pH 7.0, with 0.5% triton X-100, 10 mM EDTA, 0.5 mM of X-gluc, 0.5 mM potassium ferricyanide and 0.5 mM potassium ferrocyanide) and incubated at 37ºC overnight. Chlorophyll clearing was accomplished by submerging the leaves in 96% ethanol for 24 h and incubating at 37ºC. Once cleared, gus expression was quantified by counting blue loci per defined surface area. Data was analysed using generalized mixed model procedure with a Bonferroni adjustment for multiple comparisons. (SAS, Version 9.3, JMP® Pro 11.0.0; SAS Institute, Cary, NC).
Results
Bioinformatic strategy for candidate effector identification
The haustorial EST library that was used in this research was part of a larger P. triticina EST survey (Xu et al. Citation2011; GenBank accessions GR911120–GR911355) and global details about gene content can be found there. However, a detailed study of the secreted peptides within this haustorial library was not included in the study of Xu et al. (Citation2011). To summarize the results from the haustorial library, 188 non-redundant P. triticina specific sequences (118 contigs and 70 singlets) were found as determined by a BLASTn alignment of the unigenes the P. triticina genome, Race 1 BBBD. This also allowed for the determination of the full-length sequence of the putative gene.
One of the distinguishing features among effectors of filamentous fungi and oomycetes is the presence of a secretion signal peptide (SP). Identification of putative secreted proteins by SignalP 3.0 involves Hidden Markov Model (HMM) and neural network (NN) algorithms. Ideally, both scores should have a value of 1.00 for high confidence in the prediction. From the 188 ESTs, 10 predicted secreted proteins, with signal peptide length ranging from 18–24 amino acids (), were identified and validated by genomic PCR and sequencing (data not shown). Transcription was verified by qualitative reverse transcription-PCR with cDNA from PBJL germinated urediniospores and infected plants 6 days post-infection (dpi; ). Non-inoculated plants were used as a negative control. All but Pt3 were annotated as genes of race BBBD. However, Pt3 could be amplified from genomic DNA and cDNA of PBJL and was expressed in one of the six races from a recent RNA-seq study (Bruce et al. Citation2014). In order to elucidate the probability that the candidates were of repeated sequence origin, the candidates were analysed by GiRi (http://www.girinst.org/censor/index.php). None were positive for fungal repetitive sequences. The secreted peptide group was then subjected to nuclear localization signal (NLS) screening and no evidence was found for an NLS.
BLAST2GO functional annotation for the 10 effector candidates revealed that half of the candidates had no annotation (). Candidate Pt58 had similarity to a cell wall glucanase and Pt63 had homology to a hypothetical protein from the basidiomycete Schizophyllum commune. Pt68 was similar to a superoxide dismutase and candidate Pt69 was similar to a predicted protein from the ascomycete Botryotinia fuckeliana. Pt70 was homologous to an HESP-379-like protein from the rusts M. medusae and M. lini. The majority of the reported Avr genes from filamentous microorganisms are small secreted proteins and are often cysteine-rich (Stergiopoulos & De Wit Citation2009). In this Pt secreted peptide group, four proteins have more than three cysteine residues: Pt3 five, Pt51 nine, Pt68 five and Pt71 12 cysteine residues (). Godfrey et al. (Citation2010) identified a potential N-terminal motif, Y/F/WxC, in barley powdery mildew (Blumeria graminis f. sp. hordei) effectors, though no functional analysis has been provided. This motif is present in Pt51 at position 89, and Pt71 at position 163. No other motifs were identified in the Pt predicted secreted peptides.
Table 2. Predicted secreted proteins from a cDNA haustorial library of P. triticina race PBJL were compared with the transcriptomes of six other races (Bruce et al. Citation2014) for presence/absence and for nucleotide differences. X indicates that a transcript of this gene was found in the six race transcriptome. An * indicates that the transcript sequence of that race has a SNP that results in an amino acid change relative to BBBD.
The 10 identified genes were used in a BLASTn query against a previously generated transcriptome for six races of P. triticina (Bruce et al. Citation2014). All of the genes were found to be expressed in at least one of the races (). Since the proteins encoded by these genes are predicted to be secreted and thus may contribute to virulence/avirulence, we compared the protein sequences of these effectors among the six Pt races to detect amino acid changes that corresponded to observed virulence patterns. However, because some of the transcript assemblies from the six race data were incomplete, we were unable to associate any of the changes to a shift in virulence for the leaf rust (Lr) resistance (R) genes represented in this collection of Pt races.
Semi-quantitative reverse transcription-pcr validation
Primers for each candidate effector gene sequence were tested in samples derived from wheat leaves (p), infected leaf tissue harvested 6 dpi (i) and in vitro germinated spores (sp; ). Quality of the cDNA was evaluated using P. triticina ß-tubulin primers. Amplification products from germinated spores and infected tissue were obtained for eight candidates Pt12, Pt27, Pt58, Pt63, Pt68, Pt69, Pt70 and Pt71. Candidates Pt3 and Pt51 were only expressed in rust-infected tissue and isolated haustorial samples (). Expression was then evaluated from leaf rust-infected leaves at 0, 1, 2, 3, 4, 5 and 6 dpi. Candidate genes expressed in both spores and infected tissue did not show any change in expression from 1 to 6 dpi. Therefore, expression was evaluated at early stages of spore germination. Expression at 30 min, 1, 2 and 3 h was evaluated and no change in the presence of expression was observed ().
Transient expression in wheat tissue
The particle gun has been used to validate potential avirulence function with success in other monocot and dicot systems (Qutob et al. Citation2002). Therefore, a protocol for using the particle gun was developed for use with leaf rust Avr candidates. ß-glucoronidase (UidA) was chosen as the reporter gene to avoid autofluorescence problems associated with reporters such as GFP. Plants were grown in vitro to induce a thin cuticle for optimal GUS expression. To determine if the construct would express effectively, detached leaves from the Lr isolines were bombarded with only UidA (GUS). All lines had similar numbers of GUS expressing cells (Fellers, data not shown).
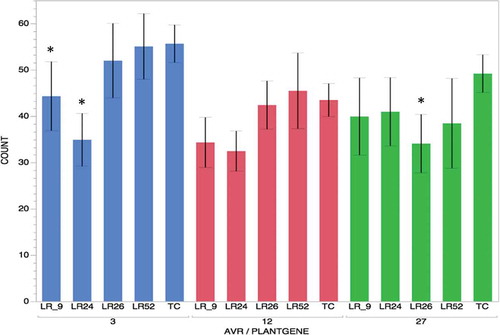
Three of the 10 candidates, Pt3, Pt12 and Pt27, were selected for bioassays based on their small size, presence of a secretion signal and cysteine content. Full-length cDNA of these genes was cloned into the plant expression vector pACH17 as BamH1 digested products and expressed under constitutive maize Ubiquitin (Ubi) promoter. Plasmids containing Ubi::UidA and Ubi::Pt candidate were co-bombarded into detached leaves from selected Lr isolines. The wheat ‘Thatcher’ was used as a control in each experiment as it is the genetic background of the isolines and susceptible to many P. triticina races (). Blue loci were manually quantified for each leaf (). Treatment combinations between Pt3 and Lr9 were found to be significantly different (P < 0.0010) from that of experimental control of Pt3 and ‘Thatcher’. ‘Treatment’ combinations between Pt3 and Lr24 were significantly different (P < 0.0001) from that of the experimental control of Pt3 and ‘Thatcher’. Treatment combinations between Pt27 and Lr26 were found to be significantly different (P < 0.0049) from that of experimental control of Pt27 and ‘Thatcher’ ( and ). Although there were reductions in number of loci in the other combinations, P values were > 0.3.
Fig. 2 (Colour online) ‘Thatcher’ isogenic lines 11 days after inoculation with P. triticina, race PBJL. PBJL induces a low infection type, rated as a fleck (;) on Lr9, Lr24, Lr26 and Lr52.
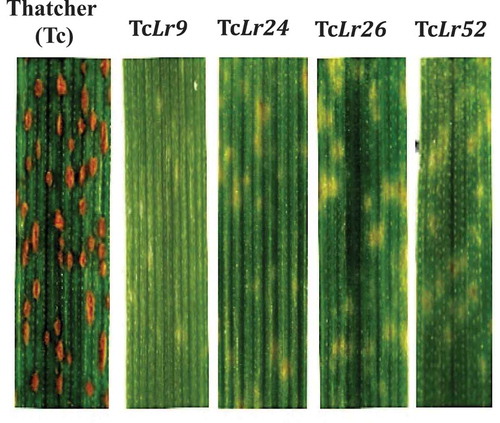
Fig. 3 (Colour online) Transient assays of P. triticina, race PBJL, avirulence candidates on the susceptible line ‘Thatcher’ versus ‘Thatcher’ isolines containing the leaf rust resistance genes Lr9, Lr24, Lr26 and Lr52. Each quadrant contains representatives of replications in the assay. Blue loci were quantified in Lr isolines and compared with ‘Thatcher’. Lines with (**) have significant reductions in blue loci at P < 0.05. Reductions are attributed to induction of the hypersensitive response.
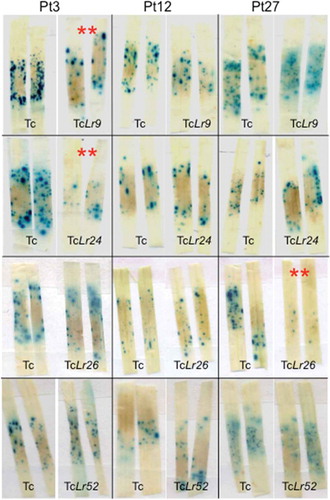
Fig. 4 (Colour online) Transient expression experiments using counts of blue loci. Pt3, Pt12 and Pt27 candidates were co-bombarded with GUS into ‘Thatcher’ isogenic lines containing leaf rust resistance genes Lr9, Lr24, Lr26 and Lr52 and ‘Thatcher’ (TC). Columns are grouped according to Pt3 (Blue), Pt12 (Red) and Pt27 (Green). Values correspond to the mean for number of spots in each interaction. * = significance at P < 0.05 compared with ‘Thatcher’ within the avirulence candidate.
Discussion
This work is technically difficult because P. triticina is an obligate biotroph and cannot be cultured in vitro. An alternative host is required for sexual crosses, which is absent in the wild in North America, and crosses are difficult to accomplish in the greenhouse. Thus, different approaches to address the questions of virulence and avirulence in P. triticina had to be used. ESTs have proved to be a useful tool to provide these answers. In this research, 188 P. triticina ESTs were derived from haustoria. Genome and EST evaluations of P. graminis f. sp. tritici (Duplessis et al. Citation2011), P. striiformis (Cantu et al. Citation2011; Zheng et al. Citation2013) and P. triticina (Xu et al. Citation2011) have shown that gene sequence homologies can vary and many of the genes are unique to each species. The genes found in this haustorial library follow a similar trend and are associated with proteins involved in metabolic processes and biological energy production. Similar findings were reported from analysis in other haustoria-forming pathogens, such as Blumeria graminis, P. striiformis and Uromyces fabae (Hahn & Mendgen Citation1997; Godfrey et al. Citation2009; Yin et al. Citation2009; Cantu et al. Citation2013; Garnica et al. Citation2013).
Previous work in flax rust (M. lini) identified 429 ESTs from haustoria. Twenty-one were predicted to be secreted proteins and four of them co-segregated with Avr loci in the fungus (Catanzariti et al. Citation2006). Of the 188 haustorial P. triticina ESTs, 10 were predicted to be secreted proteins and selected as potential Avr candidates. All the selected candidates fulfil the criteria of being secreted proteins (60–300 amino acids), except for Pt69 which is 343 amino acids long. None have a repetitive origin, are not retrotransposon sequences, nor do they have a nuclear localization signal. The Avr candidates have no significant similarities in secondary structures with proteins in the database. Many of the rust Avr genes encode small proteins with N-terminal signal peptides and are often cysteine-rich (Stergiopoulos & De Wit Citation2009; Petre et al. Citation2014). The small size and the secretion signal peptide suggest secretion from the pathogen although there is no current evidence of translocation. The cysteine residues could promote stability in the protein by forming disulphide bonds, and prevent protease degradation (Stergiopoulos & De Wit Citation2009). Two P. triticina Avr candidates, Pt3 and Pt51, were expressed only in the haustoria and are small cysteine-rich secreted proteins. The other eight candidates are expressed at an early stage of infection. Pt71 is a cysteine-rich protein with 12 cysteine residues, similar to AvrP123 from M. lini, which has 11 cysteine residues (Catanzariti et al. Citation2006). Pt68 has five cysteine residues like AvrLm6 from Leptosphaeria maculans, which has six cysteine residues (Fudal et al. Citation2007). Although six candidates have fewer than three cysteine residues or none, as in the case of Pt70, this does not negate a potential avirulence function, since some proven avirulence effectors are also poor in cysteine, such as AvrL567 and AvrM from M. lini (Catanzariti et al. Citation2006), Pwl2 from M. oryzae (Kang et al. Citation1995) and AvrLm1 from L. maculans (Gout et al. Citation2006), each with one cysteine residue.
The validation of effector function is a key task to understanding the biology of the molecular interaction in the target pathosystem, but intrinsic limitations in the biotrophic nature complicates analysis. Transient expression experiments have been used to characterize effectors in planta (Qutob et al. Citation2002; Dou et al. Citation2008; Kaneda et al. Citation2009; Kale et al. Citation2010). In cereal rusts, a non-pathogenic Pseudomonas isolate, P. fluorescens EtHAn, engineered with a Type III Secretion System (TTSS) has effectively been used to deliver effectors in wheat (Yin & Hulbert Citation2011). It relies on a Type III secretion signal (TTSS) fused to the N terminus of the candidate gene. Using this system, Upadhyaya et al. (Citation2014) identified a candidate avirulence gene from P. graminis tritici potentially interacting with stem rust resistance gene Sr22, although no proof was presented of the candidate gene interacting with Sr22. Pt27 was evaluated using the EtHAn system without a signal peptide and a nine-amino acid hemagglutinin epitope tag at its C-terminus, but an HR was not induced (G. Bakkeren, unpublished). The TTSS may interfere with proper folding of the fungal gene and with the small size of Pt27 in addition to the epitope tag, it perhaps could not be recognized by the R genes. Particle bombardment maintains the original sequence, although protein structure can be modified by the plant protein modification system. The presence of a secretion signal, as was the case in this study, may confound results.
The variability among biolistic experiments can be very high. Efficiency of the delivery and expression can vary between bombardment events (Jia et al. Citation2000; Rehmany et al. Citation2005). Particle delivery has been used to identify ATR1 (Rehmany et al. Citation2005) and ATR13 (Allen et al. Citation2004) from Hyaloperonospora arabidopsidis, Avr3a from P. infestans (Armstrong et al. Citation2005), Avrk1 and Avr10 from Blumeria graminis f. sp. hordei (Ridout et al. Citation2006) and AvrPita from M. oryzae (Jia et al. Citation2000). In this work, a statistical mixed model was used because random terms are able to be incorporated into inference from the beginning. This way, contrasts, least square means and estimates of linear combinations (interactions of Avr:Lr) are automatically reported with the correct standard errors. Generalized linear models (GLM) will not report correct P-values. Only the effects that are put in the model statement are assumed to be fixed. This allowed for the consideration for the main effects of the Avr:Lr interaction, while classifying the replication, shot and leaf(shot) in the random statement. This type of analysis was modified to use restricted maximum likelihood (REML) to estimate variance components. The value of the estimates is not changed here in relation to GLM. With this approach, Pt3 and Pt27 are associated with statistically significant reductions in blue loci for their reported isoline combinations.
The interaction of Avr gene and R gene can have significant complexity. A single Avr protein can be recognized by more than one R protein. Melampsora lini AVRL567 interacts directly with the flax L5 and L6 R proteins (Dodds et al. Citation2004). Also, the R protein can interact with multiple Avr effectors. Two effectors from P. graminis work to degrade RPG1 in barley, thus inducing a hypersensitive response (Nirmala et al. Citation2011). There has been no report of anything similar in P. triticina. In previous work, however, we have identified SNPs in one secreted peptide associated with virulence changes to several wheat R genes (Bruce et al. Citation2014). Pt27 is a small-secreted protein found in many races, but does not have a homologue in P. graminis. A sequence difference was found in race TNRJ. In the TNRJ Pt27 cDNA, a G is substituted for C at position 110, leading to an amino acid change of glycine to cysteine. Transient expression of this construct had similar reduction of blue loci as PBJL-Pt27 (data not shown). Statler (Citation2000) showed a digenic ratio of segregation for AvrLr26 and thus, Pt27 may only be part of the avirulence complex recognized by Lr26.
Three of the 10 secreted proteins were tested in transient assays to determine plant response. Pt3 induced a reduction of the number of cells expressing GUS in ‘Thatcher’ isolines containing Lr9 and Lr24. Pt27 also induced a decrease in the number of cells expressing GUS but in a line containing Lr26. It will be difficult to show direct interaction with these three R genes as each are located on chromosome fragments translocated from wheat-related species. Validation will benefit from the sequencing of more P. triticina races or by induced mutations or the analysis of a segregating wheat population.
Supplemental Material
Download MS Word (773.7 KB)Acknowledgements
The authors would like to thank Drs Barbara Valent and Ari Jumpponen for serving on Dr Segovia’s dissertation committee and providing advice throughout her training. We would also like to thank Mary Willoughby, Zach Simon, Beth Gillett-Walker, Lauren Alexander, and Sally Hermann for providing technical support.
Supplemental material
Supplemental data for this article can be accessed online here: http://dx.doi.org/10.1080/07060661.2016.1150884
Additional information
Funding
References
- Allen RL, Bittner-Eddy PD, Grenville-Briggs LJ, Meitz JC, Rehmany AP, Rose LE, Beynon JL. 2004. Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science. 306:1957–1960.
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl Acids Res. 25:3389–3402.
- Appel JA, DeWolf E, Bockus WW, Todd T 2009. Preliminary 2009 Kansas wheat disease loss estimates. Kansas Cooperative Plant Disease Survey Report. Available from: http://www.ksda.gov/includes/document_center/plant_protection/Plant%20Disease%20Reports/2009KSWheatDiseaseLossEstimates.pdf
- Armstrong MR, Whisson SC, Pritchard L, Bos JIB, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, et al. 2005. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA. 102:7766–7771.
- Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: Tools for motif discovery and searching. Nucl Acids Res. 37:W202–W208.
- Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 340:783–795.
- Bruce B, Neugebauer KA, Joly DL, Migeon P, Cuomo CA, Wang S, Akhunov E, Bakkeren G, Kolmer JA, Fellers JP. 2014. Using transcription of six Puccinia triticna races to identify the effective secretome during infection of wheat. Front Plant Sci. 4:520.
- Cantu D, Govindarajulu M, Kozik A, Wang M, Chen X, Kojima KK, Jurka J, Michelmore RW, Dubcovsky J. 2011. Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One. 6:e24230.
- Cantu D, Segovia V, MacLean D, Bayles R, Chen X, Kamoun S, Dubcovsky J, Saunders DG, Uauy C. 2013. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics. 14:270.
- Catanzariti A, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ. 2010. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant-Microbe Interact. 23:49–57.
- Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. 2006. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 18:243–256.
- Christensen A, Quail P. 1996. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transg Res. 5:213–218.
- Cokol M, Nair R, Rost B. 2000. Finding nuclear localization signals. EMBO Rep. 1:411–415.
- Conesa A, Götz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21:3674–3676.
- De Wit PJGM, Mehrabi R, Van Den Burg HA, Stergiopoulos I. 2009. Fungal effector proteins: Past, present and future. Mol Plant Pathol. 10:735–747.
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. 2004. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 16:755–768.
- Dou D, Kale SD, Wang X, Chen Y, Wang Q, Wang X, Jiang RHY, Arredondo FD, Anderson RG, Thakur PB, et al. 2008. Suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell. 20:1118–1133.
- Duplessis S, Cuomo CA, Lin YC, Aerts A, Tisserant E, Veneault-Fourrey C, Joly DL, Hacquard S, Amsele J, Cantarel BL, et al. 2011. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci USA. 108:9166–9171.
- Dyck PL, Samborski DJ. 1968. Host-parasite interactions involving two genes for leaf rust resistance in wheat. In: Findlay EW, Shepherd DW, editors. Proceedings of the 3rd International Wheat Genetics Symposium. Canberra, Australia: Australian Academy of Science; p. 245–250.
- Flor HH. 1955. Host-parasite interaction in flax rust-its genetics and other implications. Phytopathology. 45:680–685.
- Fudal I, Ross S, Gout L, Blaise F, Kuhn ML, Eckert MR, Cattolico L, Bernard-Samain S, Balesdent MH, Rouxel T. 2007. Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: Map-based cloning of AvrLm6. Mol Plant-Microbe Interact. 20:459–470.
- Garnica DP, Nemri A, Upadhyaya NM, Rathjen JP, Dodds PN. 2014. The ins and outs of rust haustoria. PLoS Pathog. 10:e1004329.
- Garnica DP, Upadhyaya NM, Dodds PN, Rathjen JP. 2013. Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS One. 8:e67150.
- Godfrey D, Böhlenius H, Pedersen C, Zhang Z, Jeppe E, Thordal-Christensen H. 2010. Powdery mildew and rust fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics. 11:317.
- Godfrey D, Zhang Z, Saalbach G, Thordal-Christensen H. 2009. A proteomics study of barley powdery mildew haustoria. Proteomics. 9:3222–3232.
- Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, Cattolico L, Balesdent MH, Rouxel T. 2006. Lost in the middle of nowhere: The AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 60:67–80.
- Hahn M, Mendgen K. 1992. Isolation by ConA binding of haustoria from different rust fungi and comparison of their surface qualities. Protoplasm. 170:95–103.
- Hahn M, Mendgen K. 1997. Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol Plant-Microbe Interact. 10:427–437.
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. Embo J. 19:4004–4014.
- Jones JDG, Dangl JL. 2006. The plant immune system. Nature. 444:323–329.
- Kale SD, Gu B, Capellut DGS, Dou D, Feldman E, Rumore A, Arredondo FD, Hanlon R, Fudal I, Rouxel T, et al. 2010. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 142:284–295.
- Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che F-S. 2009. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. Embo J. 28:926–936.
- Kang S, Sweigard JA, Valent B. 1995. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol Plant-Microbe Interact. 8:939–948.
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission, and screening of repetitive elements in Repbase. RepbaseSubmitter and censor. BMC Bioinformatics. 7:474.
- Lawrence GJ, Dodds PN, Ellis JG. 2010. Transformation of the flax rust fungus, Melampsora lini: Selection via silencing of an avirulence gene. Plant J. 61:364–369.
- McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xi XC 2010. Catalogue of gene symbols for wheat: 2010 supplement. p. 16. KOMUGI Integrated Wheat Science Database. Available from: http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2010.pdf
- McIntosh RA, Wellings CR, Park RF. 1995. Wheat rusts: An atlas of resistance genes. London, UK: Kluwer Academic.
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 15:473–497.
- Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engin. 10:1–6.
- Nirmala J, Drader T, Lawrence PK, Yin C, Hulbert S, Steber CM, Steffenson BJ, Szabo LJ, Von Wettstein D, Kleinhofs A. 2011. Concerted action of two avirulent spore effectors activates Reaction to Puccinia graminis 1 (Rpg1)-mediated cereal stem rust resistance. Proc Natl Acad Sci USA. 108:14676–14681.
- Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B. 2000. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell. 12:2019–2032.
- Ordoñez ME, Kolmer JA. 2009. Differentiation of molecular genotypes and virulence phenotypes of Puccinia triticina from common wheat in North America. Phytopathology. 99:750–758.
- Petre B, Joly DL, Duplessis S. 2014. Effector proteins of rust fungi. Front Plant Sci. 5:416.
- Qutob D, Kamoun S, Gijzen M. 2002. Expression of a Phytophthora sojae necrosis- inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32:361–373.
- Rafiqi M, Ellis JG, Victoria A, Ludowici VA, Adrienne R, Hardham AR, Dodds PN. 2012. Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Curr Opinion Plant Biol. 15:477–482.
- Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, Kamoun S, Tyler BM, Birch PRJ, Beynon JL. 2005. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 17:1839–1850.
- Ridout CJ, Skamnioti P, Porritt O, Sacristan S, Jones JDG, Brown JKM. 2006. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell. 18:2402–2414.
- Song X, Rampitsch C, Soltani B, Mauthe W, Linning R, Banks T, McCallum B, Bakkeren G. 2011. Proteome analysis of wheat leaf rust fungus, Puccinia triticina, infection structures enriched for haustoria. Proteomics. 11:944–963.
- Statler GD. 2000. Inheritance of virulence of Puccinia triticina culture X47, the F1 of the cross 71-112 x 70-1. Can J Plant Pathol. 22:276–279.
- Stergiopoulos I, De Wit PJGM. 2009. Fungal effector proteins. Annu Rev Phytopathol. 47:233–263.
- Sweigard JA, Carroll AM, Kang S, Farrall L, Chumley FG, Valent B. 1995. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell. 7:1221–1233.
- Torisky R, Fellers JP, Collins GB. 1996. A focusing device for tissue transformation with the DuPont/BioRad PDS1000 helium microprojectile system. Plant Mol Biol Rep. 14:124–133.
- Upadhyaya NM, Mago R, Staskawicz BJ, Ayliffe MA, Ellis JG, Dodds PG. 2014. A bacterial Type III secretion assay for delivery of fungal effector proteins in wheat. Mol Plant-Microbe Interact. 27:255–264.
- Valent B, Crawford MS, Weaver CG, Chumley FG. 1986. Genetic studies of fertility and pathogenicity in Magnaporthe grisea (Pyricularia oryzae). Iowa State J Res. 60:569–594.
- Webb CA, Szabo LJ, Bakkeren G, Garry C, Staples RC, Eversmeyer M, Fellers JP. 2006. Transient expression and insertional mutagenesis of Puccinia triticina using biolistics. Funct Integr Gen. 6:250–260.
- Xu J, Linning R, Fellers J, Dickinson M, Zhu W, Antonov I, Joly DL, Donaldson ME, Eilam T, Anikster Y, et al. 2011. Gene discovery in EST sequences from the wheat leaf rust fungus Puccinia triticina sexual spores, asexual spores and haustoria, compared to other rust and corn smut fungi. BMC Genomics. 12:161.
- Yin C, Chen X, Wang X, Han Q, Kang Z, Hulbert SH. 2009. Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. BMC Genomics. 10:626.
- Yin C, Hulbert S. 2011. Prospects for functional analysis of effectors from cereal rust fungi. Euphytica. 179:57–67.
- Zheng W, Huang L, Huang J, Wang X, Chen X, Zhao J, Guo J, Zhuang H, Qiu C, Liu J, et al. 2013. High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat Comm. 4:2673.
