Abstract
A severe leaf spot disease was observed on pineapple (Ananas comosus) in Fujian Province of China in July 2014. A fungus was isolated from diseased leaves showing typical symptoms of leaf spot. The pathogenicity of the isolated fungus was tested and Koch’s postulates were confirmed by re-isolation of the fungus from artificially inoculated leaves. On the basis of morphological characteristics and sequence analysis of the internal transcribed spacer (ITS) and glyceraldehyde-3-phosphate dehydrogenase (GPD) regions, the pathogen was identified as Curvularia clavata Jain. This is the first report of leaf spot of pineapple caused by C. clavata in China.
Résumé
Un cas grave de tache des feuilles a été observé sur les ananas (Ananas comosus) dans la province du Fujian, en Chine, en juillet 2014. Un champignon a été isolé à partir de feuilles infectées affichant les symptômes typiques de la tache des feuilles. La pathogénicité du champignon isolé a été testée et les postulats de Koch ont été confirmés par une nouvelle isolation du champignon inoculé artificiellement à des feuilles. Sur la base des caractéristiques morphologiques et de l’analyse de la séquence de l’espaceur transcrit interne (ITS) et des régions glycéraldéhyde-3-phosphate déshydrogénase (GPD), l’agent pathogène a été identifié en tant que Curvularia clavata Jain. Il s’agit de la première mention de la tache des feuilles sur l’ananas causée par C. clavata en Chine.
Introduction
Pineapple (Ananas comosus) is the most economically important plant in the Bromeliaceae family, producing edible multiple fruits consisting of coalesced berries. Pineapple is consumed fresh, cooked, juiced or preserved, and is found in a wide array of cuisines in many countries. It is also the only source of bromelain used in the pharmaceutical market (Moyle et al. Citation2005).
A leaf spot disease of pineapple with unknown cause became the main disease limiting pineapple production in 2014 in Zhangzhou City of Fujian Province, China. The disease incidence on pineapple leaves varied from 35% to 58% in different fields. The objective of this study was to determine the causal agent and identify the pathogen based on morphological characteristics, pathogenicity tests and molecular biological techniques.
Materials and methods
Sampling and isolation
To isolate the causal pathogen, diseased leaves were collected in July 2014 from six fields in Zhangzhou City of Fujian Province, China (117.62 E, 24.13 N). The pathogen was isolated from typical necrotic spots as follows: leaves with symptoms were washed under running water, lesion tissue (5 × 5 mm) was removed from the margin between symptomatic and healthy tissue, surface-sterilized in 0.1% mercuric chloride for 45 s, washed three to four times in sterile distilled water, and plated on potato dextrose agar (PDA) supplemented with 100 μg mL−1 rifampicin to inhibit bacterial growth. The plates were incubated at 28°C with a 12-h photoperiod for 5 days. Isolates were purified using the hyphal tip method (hyphal tips were transferred from the margin of developing colony to fresh media), and two representative isolates (BL3 and BL7) were used for identification and pathogenicity tests. Detailed microscopic examinations of specimens (spores and mycelium) were performed using a light microscope (Olympus CX51, Japan) under 400× magnification.
Pathogenicity assay
To confirm Koch’s postulates, the pathogenicity of two isolates recovered was determined by inoculating fresh and healthy detached pineapples leaves with mycelium grown on PDA. The healthy leaves were surface-sterilized and wounded with a sterile scalpel blade at five locations. Mycelial discs (4 mm diameter) removed from a 5-day-old colony on PDA were placed on the leaf wounds. Inoculation sites were wrapped with Parafilm to prevent dehydration and to hold the mycelial discs in position. Negative controls consisted of sterile PDA discs without mycelium. Each treatment included 12 leaves and the pathogenicity test was repeated three times. All treated leaves were incubated at 28°C in a moist chamber (relative humidity above 90%) with a 12-h photoperiod and observed for symptom appearance every 2 days. The pathogen was re-isolated from infected leaves, and the morphological and cultural characteristics of the pathogen were compared with the original fungus.
DNA extraction, PCR amplification and sequencing
To confirm the identity of the causal fungus, total DNA was extracted from fresh fungal mycelium of each representative isolate grown on PDA at 28°C with a 12-h photoperiod for 5 days using the genomic DNA kit (Tiangen, Beijing, China), following the manufacturer’s instructions. Internal transcribed spacer (ITS) and glyceraldehyde-3-phosphate dehydrogenase (GPD) were amplified with the primers ITS1/ITS4 (White et al. Citation1990) and GDF/GDR (Templeton et al. Citation1992), respectively. DNA amplification was performed in a 50 μL reaction mixture containing 25 uL 2×Taq PCR MasterMix (Tiangen, Beijing, China), 0.5 μM of each primer, 10 ng DNA template and autoclaved ddH2O to make 50 μL. PCR amplifications were performed in a C1000 thermal cycler system (Bio-RAD, USA) using the following program: an initial denaturation step at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 60 s, 30 s annealing at 55°C for ITS regions and 60°C for GPD, and extension at 72°C for 1 min, and a final extension step at 72°C for 10 min. Amplification products were examined by electrophoresis in 1.0% agarose gels and visualized under a UV transilluminator. PCR products were purified with a gel extraction kit (Tiangen, Beijing, China) and cloned into the Pgem-t easy vector (Promega, Madison, WI) according to the manufacturer’s instructions. Sequences in both the forward and reverse directions was conducted by a commercial sequencing service provider (Lifetech, Shanghia, China).
Sequence alignment and phylogenetic analysis
DNA sequences of tested isolates were refined using BioEdit sequence Alignment Editor (Hall Citation1999). The consensus sequences were analysed with BLAST program to identify and analyse homologous sequence with those of Curvularia species deposited in GenBank database (Altschul et al. Citation1990). Phylogenetic analysis was performed using MEGA5 with the neighbour-joining method and Tajima–Nei distance model (Tamura et al. Citation2011). The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Results and discussion
Symptoms, fungal isolation and morphological characteristics
The symptoms of leaf spot appeared as water-soaked, yellow, chlorotic or necrotic spots and greyish-brown irregular lesions (0.8 to 4.2 cm), predominantly on the leaf margin and main veins (Fig. la). Representative isolates of the causal fungus isolated from the diseased leaves and grown on PDA, produced whitish mycelia, which later became grey to dark grey (). Under microscopic examination, conidia were produced singly, and were clavate, straight or occasionally slightly curved, measuring 19 to 32 × 7.5 to 11.5 μm (n = 100), with three conspicuous septae. The basal cell was pale brown, and the central two cells were darker than the basal and apical cells. The apical cell was short and rounded, and the third cell from the base was larger and darker than the others (Fig. lc). The morphological and cultural characteristics of the isolates matched the descriptions of Curvularia clavata Jain (Mandokhot & Basu Chaudhary Citation1972).
Pathogenicity
Symptoms of leaf spot were observed in all of the inoculated leaves 7 days after inoculation, and necrotic spots similar to field symptoms were observed on wounded inoculated leaves (), while negative controls remained asymptomatic. Koch’s postulates were confirmed by re-isolation from the lesions of inoculated leaves, and colonies were recovered with the same features as those of the inoculated isolates. The results of the three repeated experiments were identical.
Molecular identification
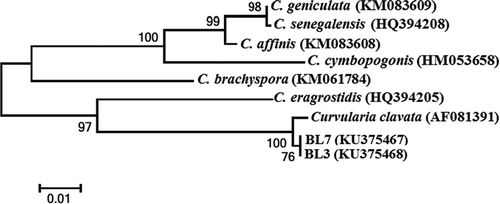
The amplified 562 bp of ITS sequence and 560 bp of GPD gene were sequenced and deposited in GenBank. The sequence was analysed using the BLAST sequence alignment algorithms from the NCBI website (www.ncbi.nlm.nih.gov). Comparison of the ITS consensus sequences of the representative isolates (accession no. KP7009559 for BL3 and KP692788 for BL7, respectively) with the GenBank database showed that the sequences of both isolates were identical and had 100% similarity with C. clavata (JQ730852) and 99% similarity with Curvularia eragrostidis (Henn.) J.A. Mey (AF163077). The GPD sequences of BL3 and BL7 (accession no. KU375468 and KU375467, respectively) were identical and showed 100% similarity with C. clavata (AF081391). The results indicated that Curvularia isolates from leaf spot lesion of pineapple should be C. clavata. In the phylogenetic tree based on GPD sequences, the representative isolate BL3 (KU375468) and BL7 (KU375467) were placed within a monophyletic clade comprising reference isolates of C. clavata (AF081391) with 100% bootstrap value (). These groupings support the identification of the representative isolates as C. clavata. Based on morphological characteristics, molecular data (sequences similarities) and pathogenicity to the host plant, the fungus causing leaf spot of pineapple in China was identified as C. clavata.
Fig. 1 (Colour online) Symptoms and morphological characteristics of the leaf spot disease of pineapple (Ananas comosus) caused by Curvularia clavata. a, Typical symptoms on naturally infected leaves, spotting and blight. b, Purified colony of Curvularia clavata isolate grown on potato dextrose agar at 28°C with a 12-h photoperiod for 7 days. c, Microscope views of spore morphology showing conidia (scale bar 20 μm). d, Detached pineapple leaves exhibiting necrotic spots and blight 7 days after inoculation with Curvularia clavata isolate BL7.
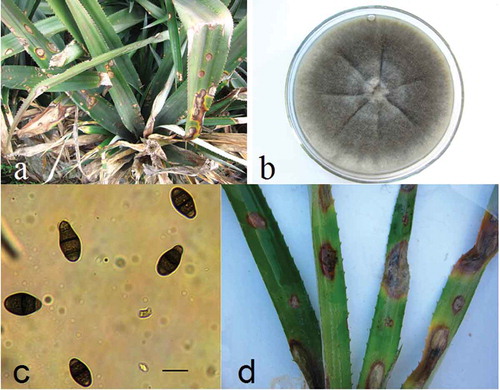
Fig. 2 Phylogenetic tree constructed by the neighbour-joining method comparing the GPD sequences of representative isolates BL3 and BL7 with sequences of related species. GenBank accessions are shown next to each isolate. Numbers on branches are the frequency with which a cluster appears in a bootstrap test of 1000 runs.
Leaf spot is one of the most destructive diseases of pineapple and causes substantial economic losses in many countries. Several fungi have been previously reported to cause leaf spot on pineapple, including C. eragrostidis (Henn) J.A. Meyer, Colletotrichum ananas Garud and Chalara paradoxa (De Seyn) Sacc (Rohrbach & Apt Citation1993), but not C. clavata. We therefore propose to include C. clavata as one of the causal pathogens of pineapple leaf spot disease in China.
The ITS region has been useful for discriminating fungi at the species level, and has been used extensively for identification and phylogenetic analysis (Gardes & Bruns Citation1993). Based on the analysis of ITS sequences, we found that C. clavata is close to C. eragrostidis. In order to confirm the identification, we included GPD sequences to discriminate our representative isolates in this study. According to our results, it is clear that using ITS sequences alone may not be adequate in distinguishing between C. clavata and C. eragrostidis, but GPD sequence as a complementary barcode to the ITS allowed differentiation between these two species.
Curvularia clavata has been reported previously as a leaf spot pathogen of many plants, such as radix curcumae (Curcuma wenyujin), physic nut (Jatropha curcas L) and maize (Zea mays) (Mandokhot & Basu Chaudhary Citation1972; Chen et al. Citation2013; Narmadhavathy et al. Citation2013), but there are no earlier reports of leaf spot caused by C. clavata on pineapple. To the best of our knowledge, this is the first report of leaf spot of pineapple caused by C. clavata in China. Such information will assist in developing biological and chemical management strategies for controlling this disease.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Chen XY, Feng JD, Su Z, Huang X. 2013. First report of Curvularia leaf blight on Curcuma wenyujin caused by Curvularia clavata in China. Plant Dis. 97:138.
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol. 2:113–118.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Mandokhot AM, Basu Chaudhary KC. 1972. A new leaf spot of maize incited by Curuvlaria clavata. Neth J Plant Pathol. 78:65–68.
- Moyle M, David J, Ripi FJ, Crowe M, Botella JR. 2005. Developing pineapple fruit has a small transcriptome dominated by metallothionein. J Exp Bot. 56:101–112.
- Narmadhavathy S, Vanitha S, Karthikeyan G, Raguchander T, Ramjegathesh R. 2013. First report of leaf blight of physic nut caused by Curvularia clavata in India. J Plant Pathol. 95:659.
- Rohrbach KG, Apt WJ. 1993. Disease of pineapple (Ananas comosus (L.) Merr.): common names of plant diseases. [Internet]; [cited 1993 May 12]. Available from: http://www.apsnet.org/publications/commonnames/pages/pineapple.asp
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Templeton MD, Rikkerink EHA, Solon SL, Crowhurst RN. 1992. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene. 122:225–230.
- White TJ, Bruns TD, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; p. 315–322.
