Abstract
The consumption of fresh young Korean ginseng roots in salads has recently increased in Korea. In July 2014, black root rot was observed on ginseng roots grown under hydroponic conditions in Ulsan, South Korea. Diseased roots first developed red lesions, which eventually became black. The typical symptoms included yellowing of the plants, stunting and wilting, reddish-brown lenticels, and black lesions on the roots. Sequencing of the internal transcribed spacer rRNA gene region and phylogenetic analysis confirmed that the isolates were Thielaviopsis basicola. Koch’s postulates were supported by pathogenicity tests conducted on healthy plants. Based on morphological characteristics, pathogenicity tests on host plants, and molecular analysis, the causal fungus was identified as T. basicola. This is the first report of black root rot caused by T. basicola on Korean ginseng. The recent occurrence of the disease indicates that the fungus poses a potential threat to fresh ginseng production in Korea.
Résumé
En Corée, la consommation de jeunes racines de ginseng dans les salades a récemment augmenté. En juillet 2014, on a observé du pourridié noir sur les racines de ginseng cultivé en hydroponie à Ulsan, en Corée du Sud. Les racines infectées présentaient des lésions rouges qui, éventuellement, noircissaient. Les symptômes typiques incluaient le jaunissement des plants, le rabougrissement et le flétrissement ainsi que des lenticelles brun-rouge et des lésions noires sur les racines. Le séquençage de la région de l’espaceur transcrit interne de l’ARNr et l’analyse phylogénétique ont confirmé que les isolats correspondaient à Thielaviopsis basicola. Les postulats de Koch ont été vérifiés par des tests de pathogénicité menés sur des plants sains. En se basant sur les caractéristiques morphologiques, sur les tests de pathogénicités menés sur les plants hôtes et sur l’analyse moléculaire, le champignon causal a été identifié en tant que T. basicola. Il s’agit de la première mention du pourridié noir causé par T. basicola sur le ginseng coréen. Cette récente incidence de la maladie indique que le champignon constitue une menace potentielle pour la production de ginseng frais en Corée.
Introduction
Ginseng (Panax L.), a member of the family Araliaceae, is one of the most important medicinal plants grown in Asia, and is also cultivated in Eastern Asia and in North America. The most common species are Korean ginseng (Panax ginseng), Japanese ginseng (Panax japonicus) and American ginseng (Panax quinquefolius). South Korea, China, Canada and the USA are the world’s largest producers of ginseng in international commerce (Evans Citation1985). Historically, Korea has been the largest producer and China the largest consumer of ginseng (Baeg & So Citation2013). Korean ginseng is popular due to its perceived benefits, including aphrodisiac and stimulant qualities, and as a cure for sexual dysfunction in men. Ginseng is available in several forms, including fresh and preserved roots in brine or in brandy. It is also available as dried whole or sliced roots, in capsule form, candied, and as chewing gum. Recently, the consumption of fresh young ginseng root in salad form has increased in Korea.
In July 2014, black root rot was observed on young roots of Korean ginseng grown for fresh salad consumption in hydroponic systems in Ulsan, South Korea. The symptoms included yellowing of the plants, stunting and wilting, reddish-brown lenticels, and black lesions on the roots. Diseased root tissue was first seen as red lesions, which eventually became black.
Here, we present the first report of black root rot caused by Thielaviopsis basicola on Korean ginseng in Korea based on distinguishing morphological features, pathogenicity tests and molecular identification using the internal transcribed spacer (ITS) rRNA gene sequence. The recent occurrence of the disease indicates that the fungus could pose a potential threat to fresh ginseng production in Korea.
Materials and methods
Fungal isolation and microscopic characterization
Fungal isolation was performed as described previously (Kwon et al. Citation2015). Briefly, root pieces (3–5 mm) taken from the margins of lesions, including both symptomatic and healthy portions of root tissue, were surface-disinfested by immersion in 1% sodium hypochlorite solution for 30 s and followed by three washes in sterile distilled water and dried on sterilized filter paper. The root pieces were placed on water agar (WA; 15 g agar in 1 L distilled water) and incubated at 25°C. Mycelial tips of the fungal isolates growing on WA were transferred to potato dextrose agar (PDA, Difco). The fresh culture of the fungus grown on PDA was examined by light microscopy (Axioplan; Carl Zeiss, Jena, Germany) under 200× magnification.
Pathogenicity tests
For pathogenicity testing, the inoculation techniques were performed as described previously with some modifications (Kwon et al. Citation2015). Young healthy roots of Korean ginseng were washed with sterile distilled water, wiped with 70% ethanol and dried. A spore suspension (106 spores mL−1) of fungal isolate MHGNU F115 was obtained by rinsing the surface of the PDA cultures with sterile distilled water. Five roots were soaked in the suspension for 5 min (Kwon et al. Citation2015). Five additional roots were soaked in sterile distilled water as a control treatment. All roots were kept in a plastic bag (20 × 22 × 15 cm) containing sterile moistened paper towels and incubated at 25°C for 7 days.
Molecular identification and phylogenetic analysis
To confirm the identity of the causal fungus, the complete ITS rRNA gene region of the two fungal isolates was amplified using the following primers: ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al. Citation1990). Total DNA was extracted using an Exgene Plant-Fungal SV Mini Kit (GeneAll Biotechnology Co., Seoul, South Korea), following the manufacturer’s instructions. The polymerase chain reaction (PCR) mixture contained 5 U of Taq polymerase (Takara, Tokyo, Japan), 1× PCR buffer, 0.2 mM of each dNTP, 5 pmol of each primer, and approximately 10 ng of fungal genomic DNA with the total volume adjusted to 50 μL with sterile water. PCR was performed using an Astec PC 802 thermal cycler (Fukuoka, Japan) with the following thermal profile: 98°C for 2 min, followed by 30 cycles of 98°C for 30 s, 60°C for 30 s and 70°C for 30 s, and a final extension step at 72°C for 4 min. The amplified products were separated by electrophoresis on a 0.8% agarose gel in 1 × Tris-borate-EDTA buffer at 50 V for 20 min. The amplicons were extracted after electrophoresis using a gel extraction kit (GeneAll), and the purified PCR products were ligated into the pGEM-T Easy Vector (Promega, Madison, WI, USA), following the manufacturer’s instructions. The ligated products were transformed into competent Escherichia coli DH5α cells by heat shock. The transformed cells were selected on a Luria–Bertani agar plate supplemented with 50 μg mL−1 ampicillin and 25 μg mL−1 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Duchefa Direct, St. Louis, MO, USA). Plasmid DNA from white transformed colonies was extracted and purified using a plasmid extraction kit (GeneAll) and checked for the expected insert size by EcoRI (Takara) digestion and visualization using gel electrophoresis and ethidium bromide staining. The resulting plasmids (pJY90 and pJY91) that contained an insert of the expected size were withheld, and the insert was sequenced in both directions using the primers M13F and M13R at Macrogen Services (Daejeon, South Korea). The resulting 556 bp of ITS rDNA sequences have been deposited in GenBank (accession nos KT696597 of MHGNU F115 and KT696598 of MHGNU F116). The sequences were analysed using the Basic Local Alignment Search Tool (BLAST) program (http://www.ncbi.nlm.nih.gov/).
A phylogenetic analysis was performed using MEGA ver. 4.1 (http://www.megasoftware.net/mega4/mega41.html) employing the neighbour-joining method and the Tajima–Nei distance model (Tamura et al. Citation2007). Multiple sequence alignment of the ITS rDNA region was performed using ClustalW. Bootstrap analysis was based on 1000 neighbour-joining data sets. Previously published ITS sequences of Thielaviopsis species were included as a reference.
Results and discussion
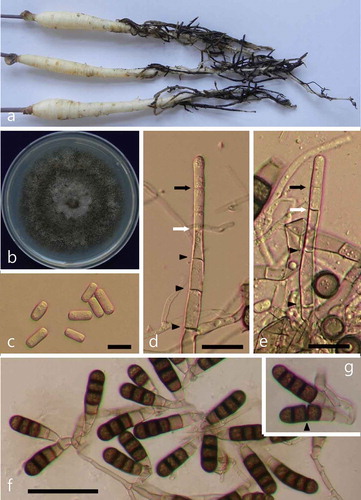
The symptoms on ginseng plants included yellowing, stunting and wilting, reddish-brown lenticels, and black lesions on the roots (). Two fungal isolates (MHGNU F115 and F116) were obtained from the diseased ginseng samples, and their colonies appeared grey-brown to black on PDA (). On microscopic examination of mycelia from the PDA cultures, two types of spores were identified. We observed single-celled hyaline endospores that were cylindrical and measured 10–15 × 3–7 μm (). Upon closer inspection, the endospores arose well within the phialide, and were fully delimited before they reached the phialide tip (–e). The aleuriospores were darkly pigmented, cylindrical, contained 2–8 cells, and measured 24–55 μm with the terminal cell having a rounded apex (). The morphological features were similar to those described previously for T. basicola (CAB International Citation2013).
Fig. 1 (Colour online) Symptoms of black root rot on Korean ginseng seedlings and morphological features of the pathogenic fungus T. basicola. (a) Symptoms of black root rot. (b) A fungal colony on PDA after 15 days of incubation. (c) Single-celled hyaline endospores (bar = 10 μm). (d–e) Production of endospores deep within the phialide (bar = 20 μm). The youngest septum in the process of formation (white arrow) and the tip of the phialide wall (black arrow) are indicated. Protoplasmic continuities across septa are shown by black arrowheads. (f) Numerous dark aleuriospores (bar = 50 μm). (g) Aleuriospore breaking up into individual spores (arrowhead).
In pathogenicity tests, Korean ginseng seedlings inoculated with the fungus started showing symptoms of rot 7 days after inoculation. At 12 days after inoculation, root rot symptoms developed on all inoculated ginseng seedlings, whereas those treated with sterilized distilled water were symptomless (Fig. S1). Thielaviopsis basicola was re-isolated from the lesions of the inoculated plants to satisfy Koch’s postulate. The morphological features of the fungus re-isolated from the inoculated ginseng seedlings were the same as those of the original isolates.
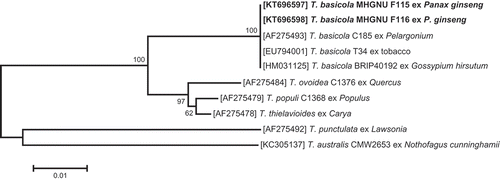
A BLAST analysis of the nucleotide sequences showed 100% identity with those of T. basicola isolates BRIP40192 (GenBank accession no. HM031125) and T34 (EU794001), which are known to infect cotton in Australia (Coumans et al. Citation2011) and tobacco in China, respectively. In the phylogenetic tree, the fungus isolated from the ginseng seedlings was placed within a clade comprising reference isolates of T. basicola ().
Fig. 2 Phylogenetic relationship based on ITS sequences, showing the closest known relatives of Thielaviopsis species. DNA sequences from the NCBI nucleotide database were aligned using ClustalW, and a phylogenetic tree was constructed using the neighbour-joining method. The numbers above the branches indicate bootstrap values. Bars indicate the number of nucleotide substitutions per site. The isolate studied in the present study is marked in bold.
Thus, on the basis of the symptoms, morphological characteristics, complete ITS sequence analysis and pathogenicity assays, the fungus was identified as T. basicola. To the best of our knowledge, this is the first report of black root rot on Korean ginseng caused by T. basicola in South Korea. The recent occurrence of the disease in ginseng indicates that black root rot could pose a serious threat to ginseng production in Korea.
Supplemental Material
Download MS Word (329 KB)Supplemental material
Supplemental data for this article can be accessed online here: http://dx.doi.org/10.1080/07060661.2016.1160956
Additional information
Funding
References
- Baeg I-H, So S-H. 2013. The world ginseng market and the ginseng (Korea). J Ginseng Res. 37:1–7.
- CAB International. 2013. Chalara elegans. Crop protection compendium. Wallingford, UK: CAB International.
- Coumans JV, Harvey J, Backhouse D, Poljak A, Raftery MJ, Nehl D, Katz ME, Pereg L. 2011. Proteomic assessment of host-associated microevolution in the fungus Thielaviopsis basicola. Environ Microbiol. 13:576–588.
- Evans BL. 1985. Ginseng: Root of Chinese-Canadian relations. Can Hist Rev. 66:1–26.
- Kwon J-H, Choi O, Kang D-W, Kim W-I KJ. 2015. The occurrence of leaf blight on Ophiopogon japonicus caused by Phyllosticta ophiopogonis in Korea. Australas Plant Dis Notes. 10:22.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 24:1596–1599.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. New York, N.Y, USA: Academic Press; p. 315–322.
