Abstract
Coconut farms located in the southern coast of Grand-Lahou in Côte d’Ivoire are severely affected by a lethal yellowing disease (CILY) associated with the group 16SrXXII-B, ‘Candidatus Phytoplasma palmicola’-related strains. Given the high prevalence of weed species on most of the farms, plants growing within and in the periphery of five selected coconut farms were assessed for the presence of the CILY phytoplasma to identify potential alternative hosts. A total of 396 plant samples belonging to 84 plant species and 35 botanical families were collected. Total DNA was extracted and tested by nested PCR with primers targeting the 16S rRNA and the translocation protein (secA) phytoplasma genes, and sequenced. Twenty samples from six plant species and five botanical families yielded PCR amplicons of the expected size, and both the secA and 16S rDNA sequences showed over 99% similarity with that of the Côte d’Ivoire lethal yellowing phytoplasma previously identified from coconut palms grown in Grand-Lahou coconut farms. Plant species from the families Poaceae (Paspalum vaginatum, Pennisetum pedicillatum), Verbenaceae (Stachytarpheta indica), Plantaginaceae (Scoparia dulcis), Phyllanthaceae (Phyllantus muellerianus) and Cyperacea (Diplacrum capitatum) were positive for the presence of the CILY phytoplasma, suggesting they may have epidemiological implications for disease spread in coconut farms in Grand-Lahou.
Résumé
Les exploitations agricoles de la côte sud de Grand-Lahou, en Côte d’Ivoire, où l’on cultive la noix de coco, sont gravement touchées par le jaunissement mortel (JM) associé au groupe 16SrXXII-B des souches apparentées à ‘Candidatus Phytoplasma palmicola’. Étant donnée la forte prévalence des mauvaises herbes sur la plupart des fermes, des plantes poussant dans cinq exploitations ciblées ou en périphérie de celles-ci ont été évaluées au regard du phytoplasme du JM afin d’identifier de possibles hôtes alternatifs. En tout, 396 échantillons de plantes appartenant à 84 espèces et 35 familles botaniques ont été collectés. L’ADN total a été extrait et testé par PCR par amorces incluses ciblant l’ARNr 16S et les gènes du phytoplasme de la protéine de translocation (secA), puis séquencé. En tout, 20 échantillons provenant de 6 espèces de plantes et de 5 familles botaniques ont fourni des amplicons de taille attendue découlant de la PCR, et les séquences de la secA ainsi que de l’ARNr 16S ont affiché une similitude de 99% avec celles du phytoplasme du jaunissement mortel de la Côte d’Ivoire préalablement identifié chez les cocotiers cultivés à Grand-Lahou. Des espèces de plantes de la famille des Poaceae (Paspalum vaginatum, Pennisetum pedicillatum), des Verbenaceae (Stachytarpheta indica), des Plantaginaceae (Scoparia dulcis), des Phyllanthaceae (Phyllantus muellerianus) et des Cyperacea (Diplacrum capitatum) se sont révélées positives quant à l’occurrence du phytoplasme du JM, ce qui suggère qu’elles peuvent avoir des implications épidémiologiques quant à la propagation de la maladie dans les exploitations de Grand-Lahou.
Introduction
Phytoplasmas are cell wall-less prokaryotic intracellular plant pathogens belonging to the class Mollicutes (Marcone Citation2014) which originated from Gram-positive bacteria. Lethal yellowing (LY)-like or Lethal Decline (LD)-like phytoplasma diseases have decimated millions of coconut groves and affected livelihoods of thousands of smallholder farm communities that depend on coconut palm trees in Africa, Central America and the Caribbean (Danyo Citation2011; Sullivan & Harrison Citation2013). Côte d’Ivoire lethal yellowing (CILY) disease is rapidly expanding into new coconut-growing villages of Grand-Lahou, a coastal commune in southern Côte d’Ivoire, and sub-prefecture of the Grand-Lahou Department in the Lagunes District. The disease has already destroyed over 400 hectares of coconut groves and continues to expand from the most CILY-affected areas (Arocha-Rosete et al. Citation2014). Moreover, the destruction of the coconut palms makes lands previously covered more exposed and therefore more prone to degradation, which adds serious environmental repercussions (Yankey et al. Citation2009).
CILY symptoms resemble those of the Cape St. Paul Wilt Disease (CSPWD) that wreaked great havoc to the coconut industry in Ghana during the last 30 years (Eziashi & Omamor Citation2010; Danyo Citation2011). The CILY phytoplasma has become a phytosanitary risk for the Ivorian coconut industry (Arocha-Rosete et al. Citation2014). Based on the analysis of the 16S rRNA gene, it was identified as a member of the subgroup 16SrXXII-B (Arocha-Rosete et al. Citation2014), officially described as ‘Candidatus Phytoplasma palmicola’-related strains (Harrison et al. Citation2014) along with the CSPWD strain from Ghana. Both CILY and CSPWD phytoplasmas have been shown to be closely related to the phytoplasma strains associated with ‘Awka Wilt’ in Nigeria and LY in Mozambique (LYM), which form the subgroup 16SrXXII-A, officially described as ‘Candidatus Phytoplasma palmicola’ (Harrison et al. Citation2014). DNA-based technology is the best approach for the detection, identification and characterization of phytoplasmas, and relies on the analyses of the 16S rRNA sequences, and their actual and/or virtual RFLP (restriction fragment length polymorphism) profiles (Lee et al. Citation1998; IRPCM Citation2004; Wei et al. Citation2007). However, the conserved nature of the 16S rRNA gene makes it difficult to discriminate among closely related strains (Duduk & Bertaccini Citation2011; Marcone Citation2014). Therefore, assays based on non-ribosomal target genes such as the secA gene are being used (Dickinson & Hodgetts 2013; Yankey et al. Citation2014; Valiunas et al. Citation2015) as a support to the 16S rRNA gene-based identification and classification system, and to distinguish among closely related phytoplasmas.
Phytoplasmas possess a double life cycle that involves replication in phloem-feeding Hemipteran insects and plant hosts. The latter involve over 1000 plant species, including crops of economic importance worldwide (Duduk & Bertaccini Citation2011). Phytoplasma vectors and alternative host plants play an active role in the development of epidemics (Lee et al. Citation2003; Weintraub & Beanland Citation2006). Alternative host plants of phytoplasma-associated diseases have been identified worldwide, and include either cultivated species or weeds. Examples of phytoplasma diseases whose secondary plant hosts have been identified include the alfalfa witches’ broom phytoplasma in Iran (Esmailzadeh-Hosseini et al. Citation2011), Napier grass stunt phytoplasma in East Africa (Arocha & Jones Citation2010), and 'bois noir' phytoplasma in grapevine in Italy (Mori et al. Citation2015).
Plant species found within surrounding crop fields have been shown to play a role in spreading phytoplasma diseases, as they may act as alternative reservoirs for the phytoplasmas (Weintraub & Beanland Citation2006) or their insect vectors (Mori et al. Citation2015). This makes the design and implementation of effective disease control strategies very difficult. So far, no alternative plant reservoirs have been identified for the CSPWD or the CILY phytoplasma strains. This paper reports on the detection and identification of the CILY phytoplasma in plant species growing in CILY-affected coconut farms of Grand-Lahou.
Materials and methods
Plant sampling
Non-coconut plant species growing within and adjacent to five coconut farms were randomly sampled during 2015. Five individual plants were collected per species in each of the CILY-affected villages: Braffedon, Palmindustrie V1, Adjadon, Yaokro and Badadon, at distances of 18 km, 26 km, 34 km, 41 km and 48 km, respectively, from Grand-Lahou. Plant samples were placed in labelled plastic bags and transported to the laboratory in a cooler with ice packs for further testing. Plant species collected were botanically identified at the University of Nangui Abrogoua (Abidjan) and the Swiss Center of Scientific Research in Côte d’Ivoire (Hutchinson & Dalziel Citation1972; Poilecot Citation1995; Aké Citation2002; Hawthorne & Jongkind Citation2006).
Polymerase chain reaction (PCR)
Leaves of the plant species collected were disinfected with 70% ethanol, and midribs were separated from leaves using a sterile scalpel. Total DNA was extracted using a CTAB small-scale protocol from 1 g of finely chopped midribs (Doyle & Doyle Citation1990), and used as a template for nested PCR assays with universal primers that target the phytoplasma 16S rRNA gene, the intergenic spacer and the beginning of the 23S rRNA. Primers included P1/P7 (Deng & Hiruki Citation1991; Schneider et al. Citation1995) for the direct PCR reaction and G813/AwkaSR (Tymon et al. Citation1998) for the nested reaction.
For all PCR reactions, 50 ng of DNA template were added to a 25 µL PCR reaction (GE Healthcare, UK). For the nested reaction, 1 µL of the 40-fold diluted direct PCR product was used. Primers SecAfor1⁄SecArev3 (direct PCR) and SecAfor5⁄SecArev2 (nested reaction) were used to amplify the secA gene that encodes the protein translocase subunit (Hodgetts et al. Citation2008). One microlitre of the direct PCR product was diluted 30-fold and used as a DNA template for the nested PCR (Dickinson & Hodgetts Citation2013). Samples were also tested with primers CoxF3 and CoxB3 that amplify the plant cytochrome oxidase (cox) gene to assess the presence of PCR inhibitors (Dickinson & Hodgetts Citation2013). Five microlitres of the PCR products were separated in a 1.5% agarose gel, and visualized with SYBR Safe (Invitrogen, USA), in a UV gel documenter (Alpha Innotech, USA).
Sequence analysis
Representative G813/AwkaSR PCR amplicons were purified on spin columns (Omega Bio-Tek, USA), cloned using p-GEMT Easy Vector (Promega, USA), and sequenced bi-directionally. The consensus G813/AwkaSR sequences were blasted (Altschul et al. Citation1990) at NCBI (http://www.ncbi.nlm.nih.gov) for phytoplasma sequence homology. Sequences obtained were aligned using Clustal W (Thompson et al. Citation1994) and phylogenetic trees were constructed using the neighbour-joining method with the program MEGA version 6.0 (Tamura et al. Citation2013) with default values and 1000 replicates for bootstrap analysis.
Results
Plant sampling
A total of 396 individual plants were collected from the five coconut-growing villages of Grand-Lahou (): Braffedon (84); Badadon (163); Palmindustrie V1 (109); Yaokro (25); and Adjadon (15), corresponding to 84 plant species and 35 botanical families, from which species from around 15 families have been previously identified as reservoirs for diverse phytoplasmas worldwide (Arocha & Jones Citation2010).
Table 1. List of plant species sampled from CILY-affected coconut farms in Grand-Lahou.
Detection of CILY phytoplasma
Phytoplasma DNA was amplified from 20 samples corresponding to six plant species and five botanical families with G813/AwkaSR primers (yielding amplicons of approximately 875 bp), from which 19 produced secA amplicons (of approximately 600 bp). CILY phytoplasma detection was consistent and reproducible from all samples that yielded both G813/AwkaSR and secA amplicons. Only three out of the 20 samples that gave positive results with the other assays also yielded P1/P7 amplifications. Cox primers yielded amplification from 387 out of the 396 plants collected, including all samples giving positive results. Nine samples did not produce any amplicons with both primer combinations G813/AwkaSR and CoxF3/B3 corresponding to Newbouldia laevis, Ixora sp. and Paullinia pinnata (Braffedon), N. laevis, Albertisia cordifolia and Cleome ciliate (Badadon), Dichapetalum heudelotii and Dissotis rotundifolia (Palmindustrie V1). This suggested the presence of possible PCR inhibitors in those samples, so they were not considered for further analyses.
Samples that gave positive results with the G813/AwkaSR assay included the following: (i) from Yaokro village: Stachytarpheta indica L. Vahl (family Verbenaceae) (); Scoparia dulcis L. (family Plantaginaceae) (); Diplacrum capitatum (Willd.) Boeckeler (family Cyperacea) (); (ii) from Adjadon village: Paspalum vaginatum Sw. (family Poaceae) (); and (iii) from Badadon, Braffedon and Palmindustrie V1 villages: Pennisetum pedicillatum Trin (family Poaceae) (), and Phyllanthus muellerianus L. (family Phyllanthaceae) ().
Sequence and phylogenetic analysis
The BLAST analysis showed that the respective G813/AwkaSR and secA sequences of the CILY phytoplasmas identified from the weeds S. indica (KU644563; KU644569), S. dulcis (KU644564; KU644570), D. capitatum (KU644565; KU644571), P. vaginatum (KU644566; KU644572), P. pedicillatum (KU644567; KU644573) and P. muellerianus (KU644568; KU644574) shared 100% sequence identity to each other and over 99% with that of the CSPWD phytoplasma strain from Ghana (Harrison et al. Citation2014) and CILY phytoplasma strains previously identified in the Grand-Lahou villages of Adjadon (KU216443; KU304343), Amanikro (KU216447), Badadon (KU216451; KU304348), Braffedon (KU216444; KU304346), Palmindustrie V1 (KU216456; KU304353) and Yaokro (KU216441; KU304337).
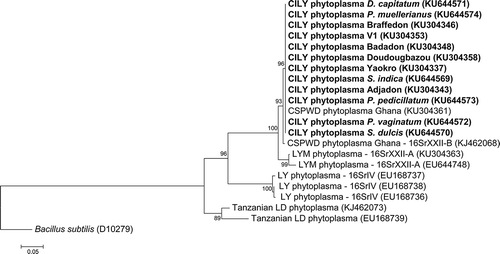
The phylogenetic trees based on the 16S rDNA () and secA () sequences showed that the CILY phytoplasma strains detected in samples from the five botanical families grouped within the same subgroup 16SrXX-B, enclosing the CILY phytoplasma strains previously reported in the CILY-affected Grand-Lahou villages, and the CSPWD phytoplasma from Ghana, which form the ‘Ca. P. palmicola’-related strains lineage, closely related to 16SrXXII-A ‘Ca. P. palmicola’ (Harrison et al. Citation2014).
Fig. 2 Phylogenetic tree based on the G813/AwkaSR sequences of the CILY phytoplasma and reference 16Sr phytoplasma groups. ‘Ca. P. sp’: ‘Candidatus Phytoplasma sp.’; CILY: Côte d’Ivoire lethal yellowing; CSPWD: Cape St. Paul Wil Disease; H. crudus: Haplaxius crudus; V1: Palmindustrie V1; LYM: Lethal Yellowing – Mozambique; LD: Lethal Decline. Ribosomal group/subgroup when available are reported before the GenBank accession numbers in parentheses.
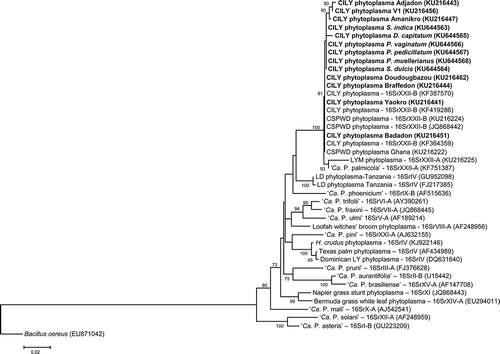
Fig. 3 Phylogenetic tree based on the secA sequences of the CILY phytoplasma and selected 16Sr phytoplasma groups. CILY: Côte d’Ivoire lethal yellowing; V1: Palmindustrie V1; CSPWD: Cape St. Paul Wilt; LYM: Lethal Yellowing – Mozambique; LD: Lethal Decline. Ribosomal group/subgroup when available are reported before the GenBank accession numbers in parentheses.
Discussion
Among the 35 botanical families identified in the natural vegetation present between rows and beneath the coconut tree canopies in the farms of the five villages of Grand-Lahou, the highest number of plant species collected belonged to the families Poaceae (45) and Fabaceae (45), followed by the families Rubiaceae (35), Asteraceae (30), Malvaceace (25) and Apocynaceae (20).
Few plant families have been reported as hosts of coconut lethal yellowing phytoplasmas. Asteraceae species such as Emelia forsbegii and Synedrella nodiflora have been identified as secondary hosts for the provisional candidate species ‘Ca. P. palmae’ (group 16SrIV) in Jamaica (Brown et al. Citation2008). Palm species, such as the African fan palm (Borassus aethiopum) and oil palm (Elaeis guineensis) from the family Arecaceae, to which coconut palms belong, have been recently reported as alternative hosts of the group 16SrXX-A ‘Ca. P. palmicola’ in Mozambique (Bila et al. Citation2015).
Plant species from five botanical families, namely Poaceae, Verbenaceae, Plantaginaceae, Phyllanthaceae and Cyperacea tested positive for the CILY phytoplasma with primers G813/AwkaSR and secA. With primers P1/P7, only three out of 20 samples positive for the CILY phytoplasma yielded amplification. This may be attributed to the well-known low titre and irregular distribution of phytoplasmas in the phloem of the infected plants (Lee et al. Citation1998).
The family Poaceae (Gramineae) has the largest reported number of plant species associated with phytoplasmas worldwide, and includes plants such as rice, wheat, barley, oats, bamboo, sorghum, sugarcane, maize, most millet, and a large range of grasses like Bermuda grass (Arocha & Jones Citation2010). Poaceae species that gave positive results for CILY phytoplasma included P. vaginatum from the Adjadon village, and P. pedicillatum from the villages of Badadon, Braffedon and Palmindustrie V1. A very well-known phytoplasma associated with Pennisetum species (from the Poaceae family) is the Napier grass stunt phytoplasma, which causes a severe stunt disease on Napier grass (P. purpureum Schumach) in Ethiopia (group 16SrIII, ‘Ca. P. pruni’) (Arocha et al. Citation2009), and in Uganda and Kenya (group 16SrXI, ‘Ca. P. oryzae’) (Arocha & Jones Citation2010). Poaceae conjugatum is another member of the Poaceae family that harbours a phytoplasma of group 16SrXIV, ‘Ca. P. cynodontis’ in Singapore (Koh et al. Citation2008). In addition, plant species of the Poaceae family, such as Panicum maximum, Sorghum arundinaceum, Andropogon gayanus and Oryza barthii have been recorded as weed species commonly found in coconut areas of Nigeria affected by the ‘Awka Wilt’ phytoplasma (Eziashi et al. Citation2013).
From the Verbenaceae family, a 16SrI (‘Ca. P. asteris’) phytoplasma was found in Verbena x hybrida plants in Turkey (Přibylová et al. Citation2014). The lethal yellowing (LY) phytoplasma (16SrIV) was identified from Stachytarpheta jamaicensis, a weed member of the Verbenaceae, which has imposed new challenges for the vector epidemiology and management of LY in Jamaica (Brown & McLaughlin Citation2011).
Plantago lanceolata L., from the family Plantaginaceae, was identified as a host of ‘Ca. P. asteris’ in the Czech Republic (Fránová & Simková Citation2009), and as one of the preferred hosts for Hyalesthes obsoletus, insect vector of the grapevine ‘bois noir’ disease in Switzerland (Kessler et al. Citation2011). Plantago major is another species within the Plantago genus that hosts the ‘stolbur’ phytoplasma in Serbia (Josic et al. Citation2012).
Phytoplasmas from groups 16SrVI, ‘Ca. P. trifoli’ and 16SrI, ‘Ca. P. asteris’ have been identified in plant species from the Phyllanthaceae family, including species such as P. amarus L. (Samad et al. Citation2004) and P. niruri L. in India (Chaube et al. Citation2015). Cyperus rotundus L. is a member of the Cyperaceae family that has been reported as a host for the phytoplasma group 16SrII, ‘Ca. P. aurantifolia’ in Cuba (Zamora et al. Citation2015). Other species within that family such as C. difformis, Mariscus sp. and Kyllingia sp. have been identified as common plants growing in coconut fields affected by CSPWD in Ghana (Danyo Citation2011).
Although Pennisetum was found in Badadon, Braffedon and Palmindustrie V1, it was not among the plant species surveyed from the villages of Yaokro and Adjadon. On the other hand, S. indica, S. dulcis and D. capitatum were limited to Yaokro, and P. vaginatum limited to Adjadon. It is not clear if there is any environmental factor driving the differential distribution or occurrence of S. indica, S. dulcis and D. capitatum in Yaokro, or P. vaginatum in Adjadon, or P. pedicillatum in either Badadon, Braffedon or Palmindustrie V1. It is possible that these specific plant species were not available at the time of sampling, or that the sampling itself was too limited. A larger sampling size would help to include more representative plant species, and assess them for the presence of the CILY phytoplasma, and further unveil any possible role in disease epidemiology.
This study provides first world reports of S. indica, S. dulcis, D. capitatum, P. vaginatum, P. pedicillatum and P. muellerianus as phytoplasma hosts. These plant species are also being reported as secondary reservoirs of the CILY phytoplasma. The confirmations were based on more than 99% sequence identity in 16Sr RNA and secA genes when compared with CILY phytoplasma strains identified from the same CILY-affected coconut farms surveyed in Grand-Lahou. No characteristic symptoms such as yellowing, witches’ broom, phyllody, or stunting, commonly associated with phytoplasmas (Lee et al. Citation1998) were observed in any of these plant species. This suggests that asymptomatic reservoirs are present naturally for CILY phytoplasma infections, as is the case of phytoplasmas identified in sugarcane and other grasses in northern Australia (Tran-Nguyen et al. Citation2000).
Phytoplasma epidemics are greatly influenced by the number and abundance of insect vectors and also by alternative/reservoir plants (Sharon et al. Citation2005). Alternative phytoplasma plant hosts may harbour additional insect vectors capable of spreading the disease (Mori et al. Citation2015). So far, no insect vector has been identified for any of the West African phytoplasmas affecting coconut palms, and associated with a lethal yellowing-like disease, including CILY. No conclusive evidence is presented in our study that supports an epidemiological role of S. indica, S. dulcis, D. capitatum, P. vaginatum, P. pedicillatum or P. muellerianus for CILY. However, these plant species were growing within the coconut plantations confirmed as infected by the CILY phytoplasma, which was also detected from the six plant species. Therefore, these species, although not confirmed as alternative hosts for the CILY phytoplasma, may pose a threat for the spread of the CILY phytoplasma in the coconut groves of Grand-Lahou. Although many of the plant species found in coconut plantations from the East, Central and Western regions in Ghana coincide with those surveyed from coconut farms in Grand-Lahou, attempts to identify alternative hosts for the CSPWD phytoplasma in Ghana have not yet been successful (Yankey et al. Citation2009).
Phylogenetic analysis also supported the utilization of the nested PCR system based on both 16Sr RNA and secA genes as a diagnostic tool for the detection of the CILY phytoplasma. In particular, the 16Sr DNA/secA nested PCR was shown to be a robust approach for detection of the CILY phytoplasma in weed species growing within and in the periphery of coconut farms in Grand-Lahou. It is noteworthy that the CILY-affected coconut farms surveyed were very poorly weeded; in particular, the villages of Badadon, Braffedon and Palmindustrie V1 exhibited a highly diverse and vigorous vegetation. Weeds are well known sources of inoculum for phytoplasma diseases (Weintraub & Beanland Citation2006; Mirzaie et al. Citation2007; Arocha et al. Citation2009); therefore their control may greatly reduce disease pressure and prevent outbreaks.
The successful uses of secA PCR in support of the 16Sr RNA-based phytoplasma identification and classification (Hodgetts et al. Citation2008; Valiunas et al. Citation2015), and for detecting a number of phytoplasmas, including the CSPWD strain in Ghana, have been reported (Yankey et al. Citation2014). The secA phylogeny of the CILY phytoplasma identified from the alternative host plants was in good agreement with the phylogenetic grouping based on the 16S rRNA gene. Both phylogenies confirmed the distinction of three ‘Ca. Phytoplasma’ taxa based on their geographic origins: Americas (LY), East Africa (Tanzanian LD) and West Africa (Nigerian LD, CSPWD in Ghana, and CILY in Côte d’Ivoire).
The use of the 16Sr DNA/secA PCR system to confirm CILY phytoplasma-presence, and the removal of S. indica, S. dulcis, D. capitatum, P. vaginatum, P. pedicillatum and P. muellerianus, should be recommended for the CILY management strategy in Grand-Lahou. Although just identified as putative alternative hosts, the surveillance of these plant species in both CILY-affected and CILY-free coconut farms will significantly contribute to better management of CILY, and improve the livelihood of smallholder coconut farmers of Grand-Lahou.
The removal of alternative hosts for phytoplasmas combined with the use of resistant plant material has proven to be highly effective for the management of lethal yellowing-like diseases. In Jamaica, a coconut rehabilitation programme ‘Black Approach’ has been successfully employed to control LY by combining the replacement of infected trees using a locally developed resistant variety and the removal of alternative reservoirs of the LY phytoplasma (Coconut Industry Board Citation2012).
Current management of CILY in Grand-Lahou includes the removal of symptomatic coconut trees, and the weeding of the farms (every 3 months) to reduce the disease pressure (Konan Konan et al. Citation2013). However, knowing the exact plant species that harbour the CILY phytoplasma will help to manage CILY more effectively by selectively removing them from the coconut farms, while providing space and less competition for other botanical species to grow that may have more resilience to CILY.
Acknowledgements
This work was carried out with the aid of a grant from the International Development Research Centre (IDRC), Ottawa, Canada (www.idrc.ca), and with financial support from the Government of Canada, provided through Foreign Affairs, Trade and Development Canada (DFATD) (www.international.gc.ca).
Additional information
Funding
References
- Aké AL. 2002. Flore de la Côte-d’Ivoire: catalogue systématique, biogéographie et écologie. II. Conservatoire & Jardin botaniques de la Ville de Genève. Boissiera. 58:1–401.
- Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Arocha Y, Jones P. 2010. Phytoplasma diseases of the Gramineae. In: Weintraub P, Jones P, editors. Phytoplasmas: genomes, plant hosts and vectors. Oxfordshire (UK): CABI publishing; p. 170–187.
- Arocha Y, Zerfy T, Abebe G, Proud J, Hanson J, Wilson M, Jones P, Lucas J. 2009. Identification of potential vectors and alternative plant hosts for the phytoplasma associated with Napier grass stunt disease in Ethiopia. J Phytopathol. 157:126–132.
- Arocha-Rosete Y, Konan Konan JL, Diallo AH, Allou K, Scott JA. 2014. Identification and molecular characterization of the phytoplasma associated with a lethal yellowing-type disease of coconut in Côte d’Ivoire. Can J Plant Pathol. 36:141–150.
- Bila J, Högberg N, Mondjana A, Samils B. 2015. African fan palm (Borassus aethiopum) and oil palm (Elaeis guineensis) are alternate hosts of coconut lethal yellowing phytoplasma in Mozambique. Afr J Biotech. 14:3359–3367.
- Brown S, Been BO, McLaughlin WA. 2008. First report of the presence of the lethal yellowing group (16Sr IV) of phytoplasmas in the weeds Emilia fosbergii and Synedrella nodiflora in Jamaica. Plant Pathol. 57:770.
- Brown SE, McLaughlin WA. 2011. Identification of lethal yellowing group (16SrIV) of phytoplasmas in the weeds Stachytarpheta jamaicensis, Macroptilium lathyroides and cleome rutidosperma in Jamaica. Phytopathog Mollicutes. 1:27–34.
- Chaube S, Mall S, Dubey DK, Rao GP. 2015. Phyllanthus niruri L.: A new host of ‘Candidatus Phytoplasma asteris’ in India. Phytopathog Mollicutes. 5:S87–S88.
- Coconut Industry Board. 2012. 71st annual report. [Internet] [revised 2014 Apr 04; cited 2016 Apr 15]. Available from: http://www.coconutindustryboardjm.org/files/2014/04/report2012.pdf
- Danyo G. 2011. Review of scientific research into the Cape Saint Paul wilt disease (CSPWD) of coconut in Ghana. Afr J Agric Res. 6:4567–4578.
- Deng S, Hiruki C. 1991. Amplification of 16SrRNA genes from culturable and non-culturable Mollicutes. J Microbiol Meth. 14:53–61.
- Dickinson M, Hodgetts J. 2013. PCR analysis of phytoplasmas based on the secA gene. In: Dickinson M, Hodgetts J, editors. Phytoplasmas: methods and protocols. Dordrecht (UK): Humana Press; p. 205–217.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:3–15.
- Duduk B, Bertaccini A. 2011. Phytoplasma classification: taxonomy based on 16S ribosomal gene, is it enough? Phytopathog Mollicutes. 1:3–13.
- Esmailzadeh-Hosseini S, Salehi M, Mirzaie A. 2011. Alternate hosts of alfalfa witches’ broom phytoplasma and winter hosts of its vector Orosius albicinctus in Yazd-Iran. Bull Insectol. 64:S247–S248.
- Eziashi E, Omamor I. 2010. Lethal yellowing disease of the coconut palms (Cocos nucifera L.): an overview of the crises. Afr J Biotech. 9:9122–9127.
- Eziashi E, Omamor I, Aisueni N, Aisagbonhi C, Airede C, Ikuenobe C, Ataga C, Oruade-Dimaro E, Odewale J, Osagie I. 2013. Potential weeds as alternate hosts of insect vectors of the lethal yellowing disease (LYD) of coconut palms (Cocos nucifera) in Nigeria. British J Appl Sci Tech. 3:123–130.
- Fránová J, Simková M. 2009. Association of ‘Candidatus Phytoplasma asteris’ with yellowing and phyllody of Plantago lanceolata. Folia Microbiol. 54:469–472.
- Harrison N, Davis RE, Oropeza C, Helmick E, Narvaez M, Eden-Green S, Dollet M, Dickinson M. 2014. ‘Candidatus Phytoplasma palmicola’, a novel taxon associated with a lethal yellowing-type disease (LYD) of coconut (Cocos nucifera L.) in Mozambique. Int J Syst Evol Microbiol. 64:1890–1899.
- Hawthorne WD, Jongkind CCH. 2006. Woody plants of Western African Forests. A guide to the forest trees, shrubs and lianes from Senegal to Ghana. Kew (UK): Royal Botanic Gardens; p. 1023.
- Hodgetts J, Boonham N, Mumford R, Harrison NA, Dickinson M. 2008. Phytoplasma phylogenetics based on analysis of SecA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. Int J Syst Evol Microbiol. 58:1826–1837.
- Hutchinson J, Dalziel JM. 1954–1972. Flora of West Tropical Africa. Vols 1–3. Crown Agents for Oversea Governments & Administrations. London (UK); p. 828, 544, 574.
- IRPCM. 2004. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol. 54:1243–1255.
- Josic D, Pavlović S, Pivić R, Kuzmanović S, Stojanović S, Popović T, Starović M. 2012. Cultivated and wild plantain (Plantago major) as a host of stolbur phytoplasma in Serbia. J Med Plants Res. 6:284–288.
- Kessler S, Schaerer S, Delabays N, Turlings TCJ, Trivellone V, Kehrli P. 2011. Host plant preferences of Hyalesthes obsoletus, the vector of the grapevine yellows disease “bois noir”, in Switzerland. Entomol Exp Appl. 139:60–67.
- Koh L, Yap M, Yik C, Niu S, Wong S. 2008. First report of phytoplasma infection of grasses in Singapore. Plant Dis. 92:317.
- Konan Konan JL, Lekadou T, N’goran B, Kouassi N, Gbalou Y. 2013. Project FIRCA (Fonds Interprofessionnel pour la Recherche et le Conseil Agricoles)/Cocotier CNRA – COC No. 588. Etude de la maladie du cocotier identifiée dans le Department de Grand-Lahou; p. 35.
- Lee I-M, Gundersen-Rindal DE, Davis RE, Bartoszyk IM. 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. Int J Syst Evol Microbiol. 48:1153–1169.
- Lee I-M, Martini M, Bottner KD, Dane RA, Black MC, Troxclair N. 2003. Ecological implications from a molecular analysis of phytoplasmas involved in an aster yellows epidemic in various crops in Texas. Phytopathology. 93:1368–1377.
- Marcone C. 2014. Molecular biology and pathogenicity of phytoplasmas. Ann Appl Biol. 165:199–221.
- Mirzaie A, Esmailzadeh-Hosseini S, Jafari-Nodoshan A, Rahimian H. 2007. Molecular characterization and potential insect vector of a phytoplasma associated with garden beet witches? Broom in Yazd, Iran. J Phytopathol. 155:198–203.
- Mori N, Quaglino F, Tessari F, Pozzebon A, Bulgari D, Casati P, Bianco A. 2015. Investigation on “bois noir” epidemiology in north-eastern Italian vineyards through a multidisciplinary approach. Ann Appl Biol. 166:75–89.
- Poilecot P. 1995. Les Poaceae de Côte d’Ivoire. Genèsve: Conservatoire et Jardin botaniques. Boissiera, vol 50, p. 734.
- Přibylová J, Petrzik K, Fránová J, Ŝpak J. 2014. Molecular characterization of aster yellows subgroup 16SrI-B phytoplasma in Verbena x hybrida. J Phytopathol. 163:1–6.
- Samad A, Ajayakumar PV, Sattar M, Khaliq A. 2004. Little leaf of Bhumyamalaki (Phyllanthus amarus) - a new case in India. Acta Phytopathol Entomol Hungarica. 39:49–56.
- Schneider B, Seemüller E, Smart CD, Kirkpatrick BC. 1995. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas. In: Razin S, Tully JG, editors. Molecular and diagnostic procedures in mycoplasmology. San Diego (CA): Academic Press; p. 369–379.
- Sharon R, Soroker V, Wesley SD, Zahavi T, Harari A, Weintraub PG. 2005. Vitex agnus-castus is a preferred host plant for Hyalesthes obsoletus. J Chem Ecol. 31:1051–1063.
- Sullivan M, Harrison N 2013. CPHST pest datasheet for ‘Candidatus phytoplasma palmae’ and related strains. [Internet]. [revised 2013 Oct 10; cited 2016 Apr 15]. Available from: http://caps.ceris.purdue.edu
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Tran-Nguyen L, Blanche KR, Egan B, Gibb K. 2000. Diversity of phytoplasmas in northern Australian sugarcane and other grasses. Plant Pathol. 49:666–679.
- Tymon AM, Jones P, Harrison NA. 1998. Phylogenetic relationships of coconut phytoplasmas and the development of specific oligonucleotide PCR primers. Ann Appl Biol. 132:437–452.
- Valiunas D, Jomantiene R, Ivanauskas A, Urbonaite I, Sneideris D, Davis R. 2015. Molecular identification of phytoplasmas infecting diseased pine trees in the UNESCO-protected Curonian Spit of Lithuania. Forests. 6:2469–2483.
- Wei W, Davis RE, Lee IM, Zhao Y. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: Identification of ten new phytoplasma groups. Int J Syst Evol Microbiol. 57: 1855–1867.
- Weintraub P, Beanland L. 2006. Insect vectors of phytoplasmas. Annu Rev Entomol. 51:91–111.
- Yankey EN, Pilet F, Quaicoe RN, Dery SK, Dollet M, Dzogbefia VP. 2009. Search for alternate hosts of the coconut Cape Saint Paul sWilt Disease pathogen. Oléagineux Corps Gras Lipides. 16:123–126.
- Yankey EN, Swarbrick PJ, Nipah JO, Quaicoe RN, Dickinson MJ. 2014. Detection of the cape St. Paul Wilt phytoplasma in coconut palms in Ghana through the combination of visual symptoms assessment and molecular diagnosis using secA gene based assay. J Plant Pathol. 96:281–285.
- Zamora L, Contaldo N, Paltrinieri S, Quiñones M, Piñol B, Acosta K, Bertaccini A. 2015. Cyperus rotundus L., a new host species for ‘Candidatus Phytoplasma aurantifolia’ - related phytoplasmas in Cuba. Phytopathog Mollicutes. 5:42–46.

