Abstract
A new foliar disease was observed on baby lima bean (Phaseolus lunatus) in fields across western New York State, USA. The disease occurred in 10 fields with variable incidence and severity. Symptoms were initially necrotic, tan spots on leaves with red to reddish brown irregular margins that coalesced to encompass the entire leaf and cause abscission. Pycnidia were observed within the lesions. Isolations from diseased leaves yielded several pycnidial fungi, including a Didymella species. These isolates were characterized by morphology and sequencing of multiple reference genes (internal transcribed spacer (ITS), partial actin, β-tubulin (tub2), translation elongation factor 1-α (TEF), 28S rDNA large subunit (LSU), rpb2 and calmodulin). A four gene phylogeny (ITS, tub2, LSU and rpb2) showed that the isolates from baby lima bean belonged to a well-supported clade that contained the type culture of Didymella americana. Pathogenicity of the isolates on three commonly grown cultivars of baby lima bean was confirmed. Symptoms that developed on inoculated plants were similar to those observed on diseased plants in the field. This is the first report of D. americana on baby lima bean.
Résumé
Une nouvelle maladie foliaire a été observée chez les jeunes haricots de Lima (Phaseolus lunatus) dans plusieurs champs de l’ouest de l’État de New York, aux États-Unis. La maladie, dont l’incidence et la gravité variaient, est apparue dans 10 champs. Initialement, les symptômes se manifestaient sous forme de taches nécrotiques brun clair avec des bordures irrégulières variant de rouge à brun rougeâtre qui confluaient pour couvrir toute la feuille et, finalement, en causer l’abscission. Des pycnides ont été observées dans les lésions. Des isolats prélevés sur des feuilles infectées ont permis d’identifier plusieurs champignons produisant des pycnides, y compris une espèce de Didymella. Ces isolats ont été caractérisés par leur morphologie et par séquençage de plusieurs gènes de référence (espaceur transcrit interne [ITS], séquence partielle d’actine, β-tubuline [tub2], facteur d’élongation de la traduction 1-α [TEF], ARNr 28S de la grande sous-unité [LSU], rpb2 et calmoduline). Une phylogénie basée sur quatre gènes (ITS, tub2, LSU et rpb2) a montré que les isolats provenant de jeunes haricots de Lima appartenaient à un clade bien établi qui comportait la culture type de Didymella americana. La pathogénicité des isolats provenant de trois cultivars de jeunes haricots de Lima cultivés couramment a été confirmée. Les symptômes qui se sont développés sur les plants inoculés étaient semblables à ceux observés au champ chez les plants infectés. Il s’agit de la première mention de D. americana infectant les jeunes haricots de Lima.
Introduction
Lima bean (Phaseolus lunatus L.) is a warm season crop grown in several regions of the USA, mainly in Delaware and the mid-Atlantic region for processing and in the Midwest and California for dry beans (Kee et al. Citation1997). Baby lima bean production has increased exponentially in western New York State, USA, from 161 ha in 2011 to over 1000 ha in 2015. Baby lima beans are planted in early June and harvested approximately 10‒12 weeks later for processing.
A severe foliar disease was reported in baby lima bean fields in New York State within two years of the start of commercial production. Symptoms on leaves began as small necrotic spots that coalesced to cover the entire leaf. Lesions were first observed on plants within 3‒4 wk of planting and increased in severity to result in substantial defoliation (). Lesions were occasionally observed on the stems, but have not been seen on pods. Pycnidia were frequently observed in the centre of lesions on leaves and stems. Isolations from lesions collected from multiple fields in 2014 and 2015 yielded two distinct pycnidial fungi. One of these fungi was identified as Boeremia exigua var. exigua which is pathogenic on baby lima bean and plays a role in the foliar disease complex (Gorny et al. Citation2015). The other fungus was tentatively identified as a Didymella sp. using the recently described generic descriptions in the Didymellaceae (Chen et al. Citation2015).
Fig. 1 (Colour online) Foliar disease symptoms caused by Didymella americana on processing baby lima bean plants in New York State, USA.

The objective of this study was to identify the Didymella sp. associated with foliar disease of baby lima bean in New York State, and determine its pathogenicity. This information is vital for gaining a comprehensive understanding of potential inoculum sources for foliar diseases, quantifying crop losses and making management decisions.
Materials and methods
Pathogen isolation and characterization
Diseased leaves were collected from 10 baby lima bean fields in western and central New York State, USA in 2014 and 2015. Leaves were surface-sterilized in 0.08% (v/v) sodium hypochlorite for 30 s and rinsed three times in sterile distilled water. Tissue pieces (approximately 2 mm2) were excised from the lesion margins, placed on 2% (w/v) water agar (WA) and incubated at 20°C under fluorescent lighting with a 12 h photoperiod. After 1‒2 days, mycelia were transferred to a new WA plate. Hyphal tips of the resultant mycelia were then transferred after 24 h onto potato dextrose agar (PDA). For long-term preservation, isolates were stored on PDA at 4°C and as mycelia at −80°C.
Two representative isolates of Didymella sp. (14098A and 14099DD) were selected for further characterization. Isolates were transferred onto six replicate malt extract agar (MEA) and oatmeal agar (OA) plates and incubated using the conditions described by Boerema et al. (Citation2004). After 7 days, mycelial growth was measured and the mean and standard error of growth rates calculated. Gross morphology, margin type and colour (Rayner Citation1970) on the upper and lower sides of the colonies were noted. For each isolate, conidial width and length were measured for 50 randomly selected conidia. The percentage of septate conidia produced on OA was also calculated. The production of metabolite ‘E’ was tested by application of a droplet of 5 M NaOH to 7-day-old cultures growing on OA following Boerema et al. (Citation2004). Colour changes or crystal production were noted at 20 min and 2 h after application. A representative isolate was deposited in the United States Department of Agriculture – Agricultural Research Service Culture Collection as NRRL 66312 (14099DD).
Pathogenicity tests
The pathogenicity of isolate 14099DD on three baby lima bean cultivars – ‘Cypress’, ‘Maestro’ and ‘Kingston’ – was tested on glasshouse-grown plants. Conidia were obtained from 30-day-old PDA cultures grown at 20°C with a 12 h photoperiod, and prepared in a suspension of sterile distilled water at a final concentration of 3 × 105 conidia mL−1 with 0.2% Tween-20 (polyoxyethylenesorbitan monolaurate). The germination rate (%) of conidia was quantified using 30 µL of the inoculum suspension on five WA plates. After incubation at 20°C for 24 h, 40 conidia were assessed for germination on each plate from three arbitrarily selected microscope fields of view (× 200), avoiding the edges. A conidium was considered to have germinated when the germ tube was at least half the length of the conidium.
Approximately 3 mL of inoculum suspension was applied to each of eight plants of each cultivar using a hand sprayer. In addition, four plants of each cultivar received the same quantity of sterile distilled water with 0.2% (v/v) Tween-20 as uninoculated controls. Following inoculation, plants were covered with plastic bags for 5 days and placed in the glasshouse at approximately 20°C. Plants were examined for disease symptoms after 25 days. Disease incidence was defined as the number of diseased plants/total number of plants × 100. Disease severity was operationally defined as the number of diseased leaves/total number of leaves per plant × 100. Isolations (n = 18/cultivar/treatment/experiment) were conducted from lesions to identify fungi associated with the lesions as described for field samples. The experimental design was a completely randomized block with inoculation and cultivar as factors and including four replications. The entire trial was repeated and data from both trials were combined to analyse the effect of inoculation and cultivar on disease incidence and severity using generalized linear modelling (Genstat 17.1; VSN International, Hemel Hempstead, UK). Means were compared using Fisher’s protected least significant difference test (P = 0.05).
Molecular identification
Mycelia (~ 100 mg) were collected from the Didymella sp. isolates using vacuum filtration from 3-day-old cultures grown in potato dextrose broth at ~ 20°C under 12 h fluorescent light. Mycelia were ground using a Tissue Lyser II system (Qiagen Inc., Valencia, CA) for 45 s at 60 hz s−1 using two 4.5 mm tungsten carbide beads. Genomic DNA was then extracted with a DNeasy Plant Mini Kit (Qiagen Inc.). DNA was resuspended in sterile distilled water and concentrations were quantified using a NanoDrop 1,000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). DNA samples were diluted to 25 ng µL−1 and stored at −20°C.
The internal transcribed spacer, partial actin, β-tubulin (tub2), translation elongation factor 1-α (TEF), 28S rDNA large subunit (LSU), rpb2 and calmodulin gene regions were amplified for the two selected isolates. The internal transcribed spacer region was amplified using primers ITS5 (5ʹ-GGA AGT AAA AGT CGT AAC AAG-3ʹ) and ITS4 (5ʹ-TCC TCC GCT TAT TGA TAT GC-3ʹ) (White et al. Citation1990). Amplification conditions for this primer set consisted of an initial denaturation at 94°C for 5 min, followed by 29 cycles of: 94°C for 1 min, 56°C for 1 min and 72°C for 1 min, and a final elongation step of 72°C for 10 min. The partial actin gene was amplified with primer set ACT-512F (5ʹ-ATG TGC AAG GCC GGT TTC GC-3ʹ) and ACT-783R (5ʹ-TAC GAG TCC TTC TGG CCC AT-3ʹ) (Carbone & Kohn Citation1999) following the PCR protocol described by Aveskamp et al. (Citation2009). For the tub2 gene, primer set BT2F (5ʹ-GTB CAC CTY CAR ACC GGY CAR TG-3ʹ) and BT4R (5ʹ-CCR GAY TGR CCR AAR ACR AAG TTG TC-3ʹ) (Woudenberg et al. Citation2009) was used with the PCR protocol of Aveskamp et al. (Citation2009). For the TEF gene, primer set EF1-983F and EF1-1567R was used based on the protocol described by Vaghefi et al. (Citation2012). The LSU region was amplified using the primers 5.8S (5ʹ-CGC TGC GTT CTT CAT CG-3ʹ) and LR7 (5ʹ-TAC TAC CAC CAA GAT CT-3ʹ) (Vilgalys & Hester Citation1990) with an amplification protocol that included an initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 45 s, 48°C for 45 s, and 72°C for 2 min, and a final elongation step of 72°C for 10 min. The rpb2 region was amplified with primer set RPB2-5F2 (Sung et al. Citation2007) and fRPB2-7cR (Liu et al. Citation1999) using the amplification protocol described by Chen et al. (Citation2015). For the calmodulin gene, primer set CAL228F and CAL2Rd (Carbone & Kohn Citation1999; Groenewald et al. Citation2013) were used with an amplification protocol of an initial denaturation at 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and a final elongation step of 72°C for 5 min.
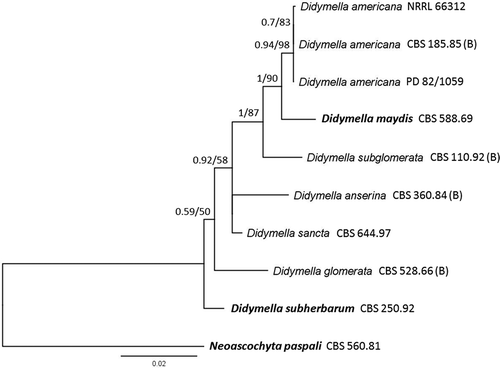
PCR reactions were conducted in a C1000 thermocycler (Bio-Rad Laboratories, Hercules, CA, USA) in a total volume of 50 µL. The PCR mixture for all primer sets with the exception of rpb2 contained 3 ng µL−1 of genomic DNA, 200 µM of each primer, 200 µM dNTP, 1.25 U Taq polymerase and 1× New England BioLabs Inc (Ipswich, MA) Standard Taq buffer. PCR products were analysed by gel electrophoresis on a 1% (w/v) agarose gel containing GelRed (Biotium, Hayward, CA) in 0.5 × TAE buffer. The PCR mixture for rpb2 was composed of 3 ng µL−1 of genomic DNA, 200 µM of each primer, 200 µM dNTP, 1.25 U New England Biolabs Q5 Polymerase and 1× New England Biolabs Q5 buffer. PCR products were prepared for sequencing using an E.Z.N.A. Cycle Pure Kit (OMEGA Bio-Tek Inc., Norcross, GA) according to the manufacturer’s instructions and a final elution volume of 40 µL. Amplicons were sequenced in both directions using the same primers for the respective genes at the Cornell University Biotechnology Resource Centre (Ithaca, NY) on an Applied Biosystems 3730xl DNA analyser. Consensus sequences were assembled by hand using pairwise alignment of the respective sequences for each gene within Geneious v. 8.1.6 (Biomatters; Kearse et al. Citation2012). Sequences from both isolates were compared with sequences from a reference strain (CBS 185.85) of Didymella americana (Morgan-Jones & J. F. White) Q. Chen & L. Cai according to Boerema et al. (Citation2004) as Peyronellaea americana, and Aveskamp et al. (Citation2009) (). Regions of variability between species were identified. Additional sequences from reference isolates were obtained from Q-bank (Bonants et al. Citation2013). Consensus sequences and reference sequences were aligned separately for each of the genes using MUSCLE (Edgar Citation2004), trimmed and edited visually. Four of the gene sequences (ITS, LSU, rpb2 and tub2) were concatenated and used for phylogenetic analysis using Bayesian inference and maximum parsimony algorithms. Neoascochyta paspali (P. R. Johnston) Q. Chen & L. Cai (syn. Phoma paspali P. R. Johnst.) CBS 560.81 was used as an outgroup for phylogenetic analyses. Bayesian inference analysis was conducted using MrBayes v3.2.2 (Ronquist & Huelsenbeck Citation2003), with the HKY85 model with gamma correction. The analysis consisted of four chains (one cold, three heated) and were run for 1 100 000 generations, with a burn in length of 100 000 generations and a random seed. Trees were sampled every 200 generations. A majority rule consensus tree was generated and labelled with consensus support as a percentage. Clades were inferred based on nodes with bootstrap values greater than or equal to 70%. Maximum likelihood phylogenies were estimated using heuristic searches in RaxML v. 7.2.8 (Stamatakis Citation2006). The GTR+GAMMA model was used with a completely random tree. Phylogenetic trees were constructed and consensus support labelled as a percentage from 1000 bootstrap replicates. Clades were inferred based on nodes with maximum likelihood bootstrap values greater than or equal to 50%.
Fig. 2 Phylogenetic relationships inferred using a Bayesian inference dendrogram between Didymella americana identified in this study (Didymella americana NRRL 66312) and other Didymella species using concatenated sequences of the ITS, tub2, LSU and rpb2 genes. CBS-KNAW Fungal Biodiversity Centre reference numbers are provided following species name. Ex-type strains are presented in bold. Species reference strains formalized by Boerema et al. (Citation2004) are marked with (B) following the CBS reference number. Numbers at branches indicate Bayesian posterior probabilities and maximum likelihood percentage bootstrap support (BPP/MLbs). Scale bar indicates the proportional genetic similarity. The dendrogram is rooted using Neoascochyta paspali CBS 560.81.
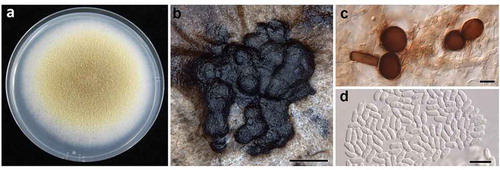
Results and discussion
Pathogen characterization
Isolates of the Didymella sp. colonies varied in colour depending upon the growth substrate. The upper surface of the colonies on PDA were honey to olivaceous buff in the centre with wide buff coloured margins (). Colonies on OA were very similar in colour to those on PDA. Colonies on PDA and OA had dense, aerial mycelia (). On MEA, the colour of colonies ranged between hazel to olivaceous buff with narrower cream to buff margins. Mycelia on MEA tended to grow down through the agar. On all media tested, the lower surface colour was olivaceous black to violaceous black. The average growth rates (± SE) on PDA, MEA and OA, were 8.4 (± 0.3), 8.1 (± 0.3) and 8.8 (± 0.1) mm d−1, respectively. The dimensions of conidia on OA were 4.5‒6.6 µm × 1.9‒3.8 µm. Pycnidia were produced profusely on the surface of the agar after 25‒30 days on all the media tested (). The production of pigmented chlamydospores was prolific at 25 days on PDA (). On all media, the conidia of both isolates were oval to oblong and mainly aseptate (< 2% septate). The dimensions of conidia on PDA were 3.1‒7.4 µm × 1.5‒3.5 µm (). Frequently, one to two oil bodies were present within the conidia. If two oil bodies were present they were positioned at opposite poles in the conidium. Assays for the production of metabolite ‘E’ were negative resulting in no colour change following application of 5 M NaOH.
Molecular identification
Amplicons sequenced from the ITS, partial actin, LSU and tub2 genes were 99‒100% identical to sequences (GenBank accession numbers FJ426972, FJ426870, GU237990 and FJ427088, respectively) of Didymella americana (CBS 185.85). Amplicons sequenced from the calmodulin gene were 98‒100% identical to the calmodulin sequence of D. americana CBS 185.85 (reference sequence obtained from Q-bank). Amplicons sequenced from the rpb2 gene were 97‒100% identical to the rpb2 sequence of D. americana CBS 185.85 (GenBank accession number KT389594). Amplicons sequenced from the TEF were 99‒100% similar among each other. However, the sequences of the TEF gene were only 40% similar to the next closest species, D. sancta CBS 644.97.
Concatenation of the four genes used in the analysis yielded a multi-locus sequence of 2488‒2546 bp in length (ITS, 357‒414 bp; tub2, 208‒209 bp; LSU, 1324‒1327 bp; rpb2, 596 bp in length, respectively). The remaining three genes produced single-locus sequences of 145‒403 bp in length (partial actin, 145‒211 bp; TEF, 236‒333 bp; calmodulin, 268‒403 bp in length). Analyses of all genes placed both isolates in a clade with the reference strain of D. americana, supporting the identification of this pathogen as Didymella americana (syn. Phoma americana Morgan-Jones & J. F. White, Peyronellaea americana (Morgan-Jones & J. F. White) Aveskamp, Gruyter & Verkley (). Sequences for NRRL 66312 have been deposited at GenBank as KT264181 (ITS), KT264182 (partial actin), KT264183 (tub2), KT264184 (TEF), KU948539 (LSU), KU948540 (rpb2) and KT264185 (calmodulin).
Pathogenicity tests
Germination rates of the conidial inoculum used for all pathogenicity trials ranged between 92 to 97%. Necrotic lesions similar to those observed on leaves in the field were observed on inoculated plants within 25 days. A small amount of disease was observed on the uninoculated plants. Isolations conducted from three of these leaves also gave rise to D. americana, suggesting a minor role of seedborne or airborne inoculum. Inoculation of baby lima bean plants resulted in significantly increased disease incidence and severity compared with the uninoculated control plants (P < 0.0001; ). Across all three cultivars, the mean disease incidence and severity was 100% and 5.5%, respectively. There was no significant difference (P > 0.05) in disease severity among the three cultivars tested. The reisolation frequency of D. americana from lesions on inoculated baby lima bean leaves was 94%.
Table 1. Effect of inoculation with Didymella americana on disease incidence (%) and severity (%) on baby lima bean leaves following inoculation in the greenhouse. Results of two experiments on three cultivars are presented as combined data.
Didymella americana has been reported in the USA on corn roots and soybean pods (Aveskamp et al. Citation2009 as Peyronellaea americana), Heterodera glycines cysts (Boerema et al. Citation2004 as Phoma americana), wheat leaves in the southern states of the USA (Morgan-Jones & White Citation1983; Boerema et al. Citation2004 as Phoma americana) and Argentina (Aveskamp et al. Citation2009 as Peyronellaea americana), sorghum in Nigeria (Aveskamp et al. Citation2009 as Peyronellaea americana) and bean (Phaseolus vulgaris) in Denmark (Aveskamp et al. Citation2009 as Peyronellaea americana). Other fungal diseases with similar symptoms reported on lima bean include B. exigua var. exigua (Gorny et al. Citation2015), pod blight caused by Diaporthe phaseolorum (Harter Citation1917), and leaf spots caused by Phyllosticta sp. and Phoma subcircinata (Halsted Citation1892).
This information is valuable as a first step towards understanding the foliar disease complex affecting baby lima bean in New York State. Necroses and defoliation associated with foliar diseases appear to be associated with multiple pathogenic pycnidial fungi. Further research is needed to quantify prevalence and temporal variations in isolation frequencies, and ultimately if crop loss is sufficient to warrant the implementation of management strategies.
Acknowledgements
This research was supported by the United States Department of Agriculture, National Institute of Food and Agriculture Hatch project NYG-625424, managed by The New York State Agricultural Experiment Station, Cornell University, Geneva, New York, a Graduate Student Extension Assistantship from Cornell University for the first author, and the Genesee Valley Regional Market Authority.
References
- Aveskamp MM, Verkley GJM, de Gruyter J, Murace MA, Perelló A, Woudenberg JHC, Groenewald JZ, Crous PW. 2009. DNA phylogeny reveals polyphyly of Phoma section, Peyronellaea and multiple taxonomic novelties. Mycologia. 101:363–382.
- Boerema GH, de Gruyter J, Noordeloos ME, Hamers MEC. 2004. Phoma identification manual: Differentiation of specific and intra-specific taxa in culture. Oxon (UK): Wellingford.
- Bonants P, Edema M, Robert V. 2013. Q-bank, a database with information for identification of plant quarantine plant pest and diseases. EPPO Bulletin. 43:211–215.
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 91:553–556.
- Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. 2015. Resolving the Phoma enigma. Stud Mycol. 82:137–217.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 32:1792–1797.
- Gorny AM, Kikkert JR, Dunn AR, Dillard HR, Smart CD, Pethybridge SJ. 2015. Tan spot of lima bean caused by Boeremia exigua var. exigua in New York State. Can J Plant Pathol. 37:523–528.
- Groenewald JZ, Nakashima C, Nishikawa J, Shin H-D, Park J-H, Jama AN, Groenewald M, Braun U, Crous PW. 2013. Species concepts in Cercospora: Spotting the weeds among the roses. Stud Mycol. 75:115–170.
- Halsted BD. 1892. Lima bean diseases. N J Agr Exp Sta 12th Ann Rep. 1891:287.
- Harter LL. 1917. Podblight of the lima bean caused by Diaporthe phaseolorum. J Agr Res. 11:473–508.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kee E, Glancey JL, Wootten TL. 1997. The lima bean: a vegetable crop for processing. HortTech. 7:119–128.
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 16:1799–1808.
- Morgan-Jones G, White JF. 1983. Studies on the genus Phoma. III. Paraphoma, a new genus to accommodate Phoma radicina. Mycotaxon. 18:57–65.
- Rayner RW. 1970. A mycological colour chart. Kew, Surrey: British Mycological Society, Commonwealth Agricultural Bureaux.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Sung G-H, Sung J-M, Hywel-Jones NL, Spatafora JW. 2007. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized congruence using a combinational bootstrap approach. Mol Phylogenet Evol. 44:1204–1223.
- Vaghefi N, Pethybridge SJ, Ford R, Nicolas ME, Crous PW, Taylor PWJ. 2012. Stagonosporopsis spp. associated with ray blight disease of Asteraceae. Aust Plant Pathol. 41:675–686.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172:4238–4246.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Spinsky TJ, White TJ, editors. PCR protocols: A guide to methods and applications. New York (NY): Academic Press; p. 315–322.
- Woudenberg JHC, Aveskamp MM, de Gruyter J, Spiers AG, Crous PW. 2009. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 22:56–62.