Abstract
Okra (Abelmoschus esculentus) is an economically important plant in China which is widely used in food, medicine and manufacturing. In September 2014, diseased samples of okra were collected in Yanqing District, Beijing City, China. Symptoms were brown to brownish black, sunken cavities on the leaves and fruits. The disease occurred on okra plants with an incidence of 35–55% in different fields. According to morphological and cultural characteristics, and sequence analysis of the internal transcribed spacer (ITS) and actin (ACT) regions, the pathogen was identified as Boeremia exigua (Desm.) Aveskamp, Gruyter & Verkley. Pathogenicity of the isolated fungus was conducted on okra fruits and on potted plants. Symptoms similar to those occurring in the field were reproduced. This is the first report of Boeremia spot of okra caused by Boeremia exigua in China.
Résumé
En Chine, l’okra (Abelmoschus esculentus) est une plante importante non seulement sur le plan économique, mais aussi pour l’alimentation, la médecine et le secteur manufacturier. En septembre 2014, des échantillons infectés d’okra ont été collectés dans le district de Yanqing de Beijing, en Chine. Les symptômes apparaissaient sous la forme de dépressions brunes à brun noir sur les feuilles et les fruits. Selon les champs, l’incidence de la maladie variait de 35 à 55%. L’agent pathogène a été identifié en tant que Boeremia exigua (Desm.) Aveskamp, Gruyter & Verkley en fonction de ses caractéristiques morphologiques et culturales ainsi que de l’analyse de la séquence des régions de l’espaceur transcrit interne (ITS) et de l’actine (ACT). Un test de pathogénicité a été mené sur le champignon isolé provenant de fruits et de plants en pot. Des symptômes semblables à ceux survenant au champ ont été reproduits. Il s’agit de la première mention de la tache causée par Boeremia exigua sur l’okra en Chine.
Introduction
Okra [Abelmoschus esculentus (L.) Moench], which is a plant of African origin belonging to the family Malvaceae, is widely cultivated in Asia, America and Europe with enormous economic value. It is used as a vegetable, a traditional medicine, as well as raw materials for paper making in China (Camciuc et al. Citation1998). In September 2014, spots on fruits and leaves were observed in Yanqing District (116°03′E, 40°32′N), Beijing. The aim of this study was to identify and describe the causal agent based on morphological characteristics and molecular approaches.
Materials and methods
Sample collection and pathogen isolation
In September 2014, diseased leaves and fruits with characteristic symptoms were collected in several fields (about 1.5 hectares representing the overall diseased area) from Yanqing District, Beijing, China. Approximately 35 samples of diseased fruits and leaves were collected from six fields, of which 10 diseased samples (four fruits and six leaves) were used for isolation. The tissues with necrotic lesions (two tissues per sample) were surface-sterilized with 75% EtOH for 30 s, and then rinsed with sterilized distilled water three times. After that, they were placed on potato dextrose agar (PDA) and incubated at 25°C with natural daylight. Subcultures were maintained by transferring 6-mm diameter mycelium plugs from the margin of older colonies to PDA, malt extract agar (MEA) and oatmeal agar (OA) plates. Cultural characteristics and conidial morphology of each colony on all three media were observed from 14-day-old cultures. Shape, length and width of 200 conidia and 60 pycnidia in total were examined by microscopy (NIKON ECLIPSE 80i, 220 V) from 20 isolated cultures.
Pathogenicity test
Pathogenicity of 20 isolated cultures was tested on fruits (15 ± 0.5 cm) and potted plants (6 weeks old) of okra ‘Jia Yuan Huang Qiu Kui’. Okra fruits were surface-sterilized by swabbing the surface with 75% EtOH. Each fruit was inoculated with three 6 mm diameter mycelial discs obtained from actively growing 7-day-old colonies on PDA. The first disc was located 4 cm from the pedicel, and the other two discs were placed at 4 cm intervals towards the base of the fruit. Nine fruits were inoculated per isolate. Potted plants were inoculated using a conidial suspension of each fungal isolate. The conidial suspension was prepared by flooding a culture on OA with 10 mL of sterilized water and gently dislodging the conidia with a brush. Each conidial suspension was adjusted to 1 × 106 conidia mL−1 and 2 mL of the conidial suspension was applied as a spray to each plant. Disease-free fruits inoculated with sterile agar plugs and healthy potted plants sprayed with sterilized water served as controls. All fruits and plants were kept in a glass cabinet (temperature 25 ± 0.5°C; RH >90%; photoperiod 12 h) for 48 h. Then the fruits and plants were transferred to a greenhouse and maintained at 25°C under natural daylight conditions for 2 weeks. The fungus was re-isolated from the infected tissues and compared with the original cultures. The experiment was repeated three times, and each replicate included three fruits and six potted plants of okra for each isolate.
Molecular identification
The total genomic DNA of all 20 isolates was extracted from pure mycelium using the Plant Genomic DNA Kit (Isolate Plant DNA Minikit, Tiangen, China) following the manufacturer’s instructions. The internal transcribed spacers (ITS) and actin (ACT) genes were amplified using primer pairs ITS1 (5ˊ-TCCGTAGGTGAACCTGCGG-3ˊ)/ITS4 (5ˊ-TCCTCCGCTTATTGATATGC-3ˊ), and ACT-512F (5ˊ-ATGTGCAAGGCCGGTTTCGC-3ˊ)/ACT-783R (5ˊ-TACGAGTCCTTCTGGCCCAT-3ˊ), respectively (de Gruyter et al. Citation2012). Polymerase chain reaction (PCR) was performed using 1.6 µL of dNTP mix (2.5 mM µL−1), 0.2 µL of Taq polymerase (5 U µL−1), 1 µL of genomic DNA (50 ng µL−1), 2 µL of polymerase buffers (10 X µL−1, TaKaRa, Japan), 1 µL of each primer (25 pmol µL−1), and total volume was adjusted to 20 µL with ddH2O, in an Applied Biosystems thermocycler (BIO-RAD, S1000 Thermal Cycler, USA). The following thermocycling pattern was used to amplify the ITS region: an initial pre-heat at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min, terminating with a final extension at 72°C for 10 min. The PCR cycle conditions for ACT gene were 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, 61°C for 30 s, 72°C for 30 s and a final elongation step at 72°C for 10 min. The PCR product was sequenced and the results were compared with those available in NCBI’s GenBank database (http://www.ncbi.nlm.nih.gov). Sequences were aligned using MEGA 5.1 software package (Tamura et al. Citation2011), and phylogenetic analysis was conducted using the Neighbour-Joining (NJ) method with a bootstrap of 1000 replicates (Saitou & Nei Citation1987) based on the internal transcribed spacers (ITS) sequence of our isolates. Botrytis cinerea served as the outgroup (GenBank accession no. AB921204).
Results and discussion
Fungal isolation and morphological characteristics
Disease symptoms on okra plants included lesions on fruits and leaves. Spots on fruits were sunken, oval to irregular, brownish black, 2.5–5 × 1–2.5 cm in size (). Eventually, the fruits senesced and were rotted. Spots on leaves were circular to irregular in shape, medium brown to greyish white in colour, up to 1.3 cm in diameter (). At the later stages, large areas of the leaves with lesions dried up.
Fig. 1 (Colour online) Symptoms of (a–b) brownish black spots on fruits, and (c) medium brown to greyish white spots on leaves of naturally infected okras in the field. Colony on (d) PDA, (e) MEA and (f) OA at 25°C for 14 days. Microscopic structures of (g) pycnidia and (h–i) conidia. Symptoms observed in pathogenicity tests (j–k) on fruits, and (l) on leaves. Bars: g = 100 μm, h-i = 20 μm.

A total of 20 colonies with the same morphological characteristics were recovered from diseased tissues, with a frequency of recovery of 100%. Colonies on PDA at 25°C after 14 days were grey-green, with white margins and aerial hyphae (). On MEA, colonies were white, smooth with woolly floccose, concolourous to grey mycelia (); on OA, colonies were irregular, greenish olive to olive grey with scanty, floccose, white mycelia (). Pycnidia were scattered, globose to irregular, 95–153 µm wide, 70–98 µm in length () (based on 60 pycnidia measurements). Conidia were hyaline, subglobose, ellipsoidal to oblong, mainly aseptate, occasionally 1-septate, 3.0 to 10.5 × 1.3 to 4.2 μm () (based on 200 spore measurements). Morphological characteristics of the pathogen were similar to original reports for Boeremia exigua (Desm.) Aves., Gruy. & Verk. (Aveskamp et al. Citation2010).
Pathogenicity test
Typical symptoms first appeared on fruits 5 days after inoculation, and developed on leaves within 7 days after inoculation. Symptoms were similar to those observed on okra plants in the field. The lesions on fruits were brownish-black, sunken (, k). The spots on leaves were brown, subcircular to irregular with greyish white centres (). The disease incidence in inoculated fruits and plants was 100%. All the control plants without inoculum showed no signs of damage. Re-isolation of the infected tissues showed morphological and cultural properties as in the original isolates.
Molecular identification
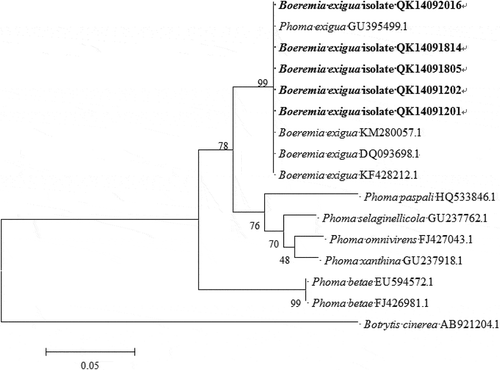
The amplicon size of ITS and ACT was c. 416 and 272 bp, respectively, from all 20 isolates. The sequences of ITS and ACT genes of the isolate QK14091201 have been submitted to NCBI database (GenBank accession no. KP942364 and KP942365, respectively). Sequence analysis of the amplified product showed 100% identity to the ITS sequence of strain ZJUB106 of Boeremia exigua isolate (KR653198.1) and 99% identity to the ACT sequence of culture-collection ICMP:16851 of Boeremia exigua (KT309240.2). A phylogenetic tree was obtained through the neighbour-joining method using the MEGA 5.1 program based on the ITS sequence of our isolates (QK14092016, QK14091814, QK14091805, QK14091202 and QK14091201). Our isolates clustered with Boeremia exigua and were clearly distinguished from other Phoma spp., including P. paspali, P. selaginellicola, P. omnivirens, P. xanthina and P. betae (), proving the identity of the isolates as B. exigua. Additionally, ITS and ACT sequences retrieved from the re-isolated fungus showed 100% identity with the data from field isolates. The results showed that Boeremia exigua was the causal agent of the disease.
Fig. 2 Phylogenetic tree obtained through the neighbour-joining method using the MEGA 5.1 program based on the internal transcribed spacers (ITS) sequence of five isolates from this study, and 10 isolates retrieved from GenBank. Bootstrap support values (%) resulting from 1000 replicates are shown at the branch points. Botrytis cinerea served as the outgroup.
Boeremia is a new genus created by Aveskamp, Gruyter and Verkley in 2010 based on multiple analyses of 206 taxa, including 159 known to Phoma and related pleosporalean genera. Boeremia is the largest genus in the Sphaeropsidales, which infects many agronomically and commercially important crops throughout the world (Koike et al. Citation2006). Boeremia exigua, which is the type species of the genus Boeremia, is considered to be a destructive pathogen (Aveskamp et al. Citation2010), causing disease on cotton (as Phoma exigua) (Koenning et al. Citation1999) and hollyhock (as Phoma exigua) (Irinyi et al. Citation2009), both members of the Malvaceae. Boeremia spot disease has only been recorded on A. esculentus in Brazil to date (as Phoma exigua) (Mendes et al. Citation1998; Farr & Rossman Citation2016). In China, B. exigua has been recorded on pepper (as Phoma exigua) (Liang Citation1991), tobacco (as Ascochyta nicotianae) (Bai Citation2003), lentil (as Ascochyta phaseolorum) (Guo Citation1997) and other plants. Abeln et al. (Citation2002) proposed to identify B. exigua not only based on phenotypic characters, but also strongly supported by molecular genetic analysis. On the basis of the research in this paper, Boeremia exigua is the first report on Abelmoschus esculentus (L.) Moench in China.
Additional information
Funding
References
- Abeln ECA, Stax AM, de Gruyter J, van der Aa HA. 2002. Genetic differentiation of Phoma exigua varieties by means of AFLP fingerprints. Mycol Res. 106:419–427.
- Aveskamp MM, de Gruyter J, Woudenberg JHC, Verkley GJM, Crous PW. 2010. Highlights of the Didymellaceae: A polyphasic approach to characterize Phoma and related pleosporalean genera. Stud Mycol. 65:1–60.
- Bai JK. 2003. Flora Fungorum Sinicorum. Vol. 17. Sphaeropsidales, Ascochyta, Septoria. Beijing (China): Science Press.
- Camciuc M, Deplagne M, Vilarem G, Gaset A. 1998. Okra—Abelmoschus esculentus (L.) Moench a crop with economic potential for set aside acreage in France. Indus Crops Prod. 7:257–264.
- de Gruyter J, van Gent-Pelzer MPE, Woudenberg JHC, van Rijswick PCJ, Meekes ETM, Crous PW, Bonants PJM. 2012. The development of a validated real-time (TaqMan) PCR for detection of Stagonosporopsis andigena and S. crystalliniformis in infected leaves of potato and tomato. Eur J Plant Pathol. 134:301–313.
- Farr DF, Rossman AY. 2016. Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA [Internet]; [ cited 2016 Mar 14]. Available from: http://nt.ars-grin.gov/fungaldatabases/.
- Guo YL. 1997. Fungal flora of the Daba mountains: imperfect fungi. Mycotaxon. 61:13–33.
- Irinyi L, Kövics GJ, Sándor E. 2009. Taxonomical re-evaluation of Phoma-like soybean pathogenic fungi. Mycol Res. 113:249–260.
- Koenning SR, Abdel Alim FF, Grand LF, Phipps PM. 1999. Stem canker of cotton caused by Phoma exigua in North Carolina and Virginia. Plant Dis. 83:1251.
- Koike ST, Subbarao KV, Verkley GJM, Fogle D, O’Neill TM. 2006. Phoma basal rot of romaine lettuce in California caused by Phoma exigua: occurrence, characterization, and control. Plant Dis. 90:1268–1275.
- Liang LZ. 1991. A new variety of Phoma exigua. Acta Micro Sin. 31:160–162.
- Mendes MAS, da Silva VL, Dianese JC, Ferreira MASV, dos Santos CEN, Gomes NE, Urben AF, Castro C. 1998. Fungos em Plants no Brasil. Brasilia, Embrapa-SPI/Embrapa-Cenargen.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
