Abstract
A severe leaf spot disease was observed in a commercial tomato greenhouse in Dalian, Liaoning Province, China in January 2014. Typical symptoms included necrotic spots with dark brown, slightly sunken, irregular lesions on the leaves and stems, leading to wilting. The causative pathogen was identified as Fusarium proliferatum (Matsush.) Nirenberg on the basis of morphological characteristics and PCR amplification and sequence analysis of five genes, namely translation elongation factor-1α (TEF), β-tubulin (BT), fumonisin biosynthetic polyketide synthase (FUM1), histone H3 (H3) and calmodulin (Cal). Pathogenicity was confirmed by fulfilling Koch’s postulates. To our knowledge, this is the first report of leaf spot disease on tomato caused by F. proliferatum in China.
Résumé
En janvier 2014, un cas grave de la tache foliaire a été observé sur les plants de tomate d’une serre commerciale à Dalian, dans la province du Liaoning, en Chine. Les symptômes typiques incluaient des taches nécrotiques affichant des lésions irrégulières brun foncé et légèrement concaves sur les feuilles et les tiges, entraînant le flétrissement des plants. En se basant sur les caractéristiques morphologiques et l’amplification par PCR ainsi que sur l’analyse de séquence de cinq gènes, dont le facteur d’élongation de la traduction-1α (FET), la ß-tubuline (BT), le gène FUM1 qui code pour la polycétide synthase impliquée dans la biosynthèse des fumonisines, l’histone H3 (H3) et la calmoduline (Cal), l’agent pathogène causal a été identifié comme étant Fusarium proliferatum (Matsush.) Nirenberg. La pathogénicité a été confirmée en remplissant les postulats de Koch. À notre connaissance, il s’agit du premier rapport de la tache foliaire chez la tomate, causée par F. proliferatum en Chine.
Introduction
Tomato (Solanum lycopersicum L.) is a major crop plant and a model system for fruit development (Consortium Citation2012). Tomato is affected by various pathogens, such as Botrytis cinerea Pers., Alternaria solani (Ellis & Martin) Sorauer and Phytophthora infestans (Mont.) de Bary, which have caused significant reductions in yield and resulted in economic losses throughout the world. Another well-known tomato pathogen is Fusarium species, which cause root rots and wilt disease. Fusarium wilt, which is a vascular wilt disease caused by F. oxysporum f. sp. lycopersici Snyder & Hansen (Fol), is highly prevalent in tomato (Abdallah et al. Citation2016). It is highly destructive and has previously caused great economic loss to tomato crops in at least 32 countries (McGovern Citation2015).
In January 2014, about 600 tomato plants with c. 50% displaying symptoms of necrotic spots and wilt were detected out of 3000 plants in a commercial tomato greenhouse in Dalian, Liaoning Province, China. Initial symptoms on infected leaves and stems were small, water-soaked, brown to black lesions. The lesions increased to 1–3 mm in diameter and became irregular, dull brown leaf spots, resulting in rapid necrosis with abundant whitish mycelia on the surface of infected tissues (). The small necrotic spots continued to enlarge, leading to softened and wilted leaves, which either fell off the plant or remained attached in the advanced stages of disease. These symptoms are similar to those manifested by diseases caused by Fusarium species (Conner et al. Citation1996; Eken et al. Citation2004).
Fig. 1 (Colour online) Leaf spot of tomato caused by F. proliferatum. (a) Necrotic lesions on tomato leaf in commercial tomato greenhouse; (b) 7-day-old colony of F. proliferatum growing on potato dextrose agar at 20°C; (c) Fully grown colony with sclerotia; (d) Macroconidia and microconidia of the fungus formed on PDA; (e), (f) Necrotic spots on tomato leaf and stem infected with F. proliferatum 4 days after artificial inoculation.

Sequence analysis of the internal transcribed spacer region (ITS) of rDNA is often used to support the morphological identification of an organism isolated from plant tissues. However, many fusaria within the Gibberella clade possess non-orthologous copies of ITS2, which can lead to incorrect phylogenetic inference (O’Donnell & Cigelnik Citation1997; O’Donnell et al. Citation1998). The translation elongation factor-1α gene (TEF) is regarded as a good tool for the identification of Fusarium species because it is highly informative at the species level and non-orthologous copies of the gene have not been detected in the genus (Geiser et al. Citation2004). The β-tubulin (BT), fumonisin biosynthetic polyketide synthase (FUM1), histone H3 (H3) and calmodulin (Cal) genes have also been used for the identification of Fusarium species. The purpose of this study was to identify the causative pathogen associated with the necrotic spots of tomato plants in Dalian through Koch’s postulates and molecular methods.
Materials and methods
Sampling and isolation
Samples consisting of infected leaves and stems with necrotic spots were collected from 24 tomato plants in a commercial tomato greenhouse in Dalian, Liaoning Province, China (c. 38º89′N, 121º26′E) where the disease was detected in January 2014. Each sample was surface-sterilized by immersion in 70% ethanol for 30 s, then in 2% NaOCl for 1.5 min followed by rinsing thrice in sterilized distilled water. Leaves were cut into 2–3 mm pieces with a sterile scalpel and then transferred onto 2% water agar dishes amended with 100 mg L−1 streptomycin sulphate. After 2 days of incubation at 20°C, agar pieces with mycelia growth from the infected tissues were transferred to potato dextrose agar (PDA) and incubated at 20°C for 7 days. The fungal colonies that appeared on PDA were then subcultured on fresh PDA plates by transferring mycelia plugs from the edge of the colonies onto the medium with a sterile needle. The fungal colonies were purified by obtaining a single conidial isolate by micromanipulation according to Leslie & Summerell (Citation2006). The morphology of the colony and conidia of a 7-day-old culture were observed with a light microscope at 400 × magnification. Thirty conidia were measured in each experiment, and the experiment was performed three times.
Pathogenicity tests
A pathogenicity test using a representative fungal culture was conducted on 2-month-old healthy plants of tomato (S. lycopersicum ‘Zaofen No. 2ʹ) grown individually in plastic pots (20 cm diameter × 14 cm height) filled with sterile soil under glasshouse conditions. Twenty-four tomato plants of the same age and condition were used in this experiment. The leaves and stems of 1 group (12 plants) were sprayed to the point of runoff with a spore suspension (2 × 105 spores mL−1) harvested from the fungus grown on PDA at 20°C in darkness for 7 days. The other group (12 plants) was used as control, and the leaves and stems of these plants were sprayed with sterile distilled water only. The plants were placed in a growth chamber set at 25°C, 100% relative humidity and a 16 h day–8 h night cycle following an initial period of 24 h in the dark. They were observed daily for symptom development. To satisfy Koch’s postulates, the causal fungus was re-isolated from the infected tissues of the inoculated plants, and its morphological characteristics were compared with those of the original isolate. The pathogenicity experiment was repeated three times.
Molecular identification
Genomic DNA was extracted from the mycelia of a colony using the CTAB method (Lee & Taylor Citation1990). Gene-specific primers used in PCR to detect the presence of the translation elongation factor-1α (TEF), β-tubulin (BT), fumonisin biosynthetic polyketide synthase (FUM1), histone H3 (H3) and calmodulin (Cal) genes in the extracted genomic DNA were taken from previous studies (Geiser et al. Citation2004; Jeon et al. Citation2013; Chang et al. Citation2015), since these five genes have specifically been used for Fusarium species identification. The PCR products were purified using the TaKaRaMiniBESTAgarose Gel DNA Extraction Kit Ver. 3.0 (TaKaRa Corporation Ltd, Dalian, China) and subjected to DNA sequencing performed by Beijing Genomic Institute (BGI, China). The DNA sequences from each of the genes were compared with the National Center for Biotechnology Information (NCBI) database for sequence identity. A phylogenetic tree was constructed by the neighbour-joining method using MEGA6 (Tamura et al. Citation2013). The robustness of the neighbour-joining tree was evaluated by 1000 bootstrap replicates.
Results and discussion
Sampling and isolation
Colonies grew rapidly when infected plant tissues were cultured on PDA, giving rise to aerial mycelia which turned cottony-white after 7 days, while the reverse side became pale yellow to orange (). About 83% isolate recovery was observed from the infected tissues. After 3 weeks, numerous white-coloured and subglobose sclerotial bodies were produced on the surface of the cultures (). The macroconidia were hyaline, slender, slightly falcate, 3–5 septa, and measured 12.0–32.5 × 2.5–4.0 µm. The microconidia were abundant, single-celled, ovoid, hyaline, aseptate, and about 4.5–10.5 × 1.0–2.5 µm in size (). The cultural and morphological characteristics of the fungus were consistent with those of Fusarium proliferatum (Matsush.) Nirenberg (Nelson et al. Citation1983; Leslie & Summerell Citation2006).
Pathogenicity tests
Small necrotic spots appeared on the leaves and stems of inoculated tomato plants 3–4 days after inoculation (, f). Initially, water-soaked lesions appeared on inoculated leaves 2 days post-inoculation and became dark brown and enlarged rapidly. Leaves and stems were softened and wilted 7 days after inoculation. In the advanced stages of the disease, infected leaves and stems became rotten and fell off the plants. All inoculated tomato plants were found to exhibit the symptoms, but no dead plants were observed. No symptoms were observed on the control tomato plants sprayed with sterile water. The fungus re-isolated from lesions of the inoculated plants was morphologically identical to that originally isolated from the diseased plants, thus fulfilling Koch’s postulates. Isolates were recovered from all the inoculated plants.
Molecular identification
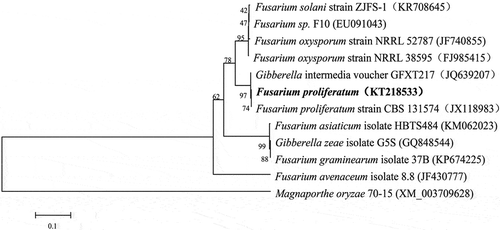
All the PCR products amplified with the five sets of F. proliferatum-specific primers showed complete sequence identity with those found in GenBank database (). The phylogenetic analysis of the TEF sequences showed that the strain (accession no. KT218533) clustered with F. proliferatum, forming a separate clade distinct from other species of Fusarium in a neighbour-joining tree (). This result further confirmed the identification obtained from morphological analysis. The data therefore indicated that F. proliferatum was the microorganism that caused the disease in the tomato plants in Dalian, China. Fusarium proliferatum is a prevalent pathogen of agriculturally important crops, and has a wide host range (Leslie & Summerell Citation2006). It has been reported to cause black point of wheat (Conner et al. Citation1996), stalk and ear rot of maize (Bailey et al. Citation2003), root rot of soybean (Chang et al. Citation2015), wilt disease of carnation (Zhang et al. Citation2013) and crown rot of gypsophila (Lee et al. Citation2011), as well as diseases of winter jujube (Zhang et al. Citation2012), citrus fruit and banana (Leslie & Summerell Citation2006), onion (Bayraktar & Dolar Citation2011) and garlic bulbs (Tonti et al. Citation2012). We have isolated only one Fusarium species from the infected tomato plants, although other investigators have recently reported the isolation of several different Fusarium species from the stems of diseased tomato plants (Imazaki & Kadota Citation2015).
Table 1. Sequence identities for the five genes found by a BLAST search in the GenBank database.
Fig. 2 Phylogenetic tree constructed with sequences of partial translation elongation factor-1α (TEF). Gene sequences show the closest known relatives of F. proliferatum, inferred by the neighbour-joining method and reference sequences retrieved from GenBank database. The per cent bootstrap support values (1000 replications; ≥70%) are shown in the branches. Bar = number of nucleotide substitutions per site.
In conclusion, this is the first report of F. proliferatum causing leaf spot disease of tomato plants in China. During the winter, cool and humid weather tends to be favourable for F. proliferatum to cause diseases on an epidemic scale in China. Fusarium proliferatum can produce toxins that are harmful to animals (Price et al. Citation1993). Therefore, this fungus could be a potential threat to tomato production and may have an impact on the health of consumers. More research is needed to better understand the biology of the pathogen, the process of disease development, and to develop tomato cultivars resistant to Fusarium leaf spot.
Additional information
Funding
References
- Abdallah RAB, Mokni-Tlili S, Nefzi A, Jabnoun-Khiareddine H, Daami-Remadi M. 2016. Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biol Control. 97:80–88.
- Bailey KL, Gossen BD, Gugel RK, Morrall RAA. 2003. Diseases of field crops in Canada. Saskatoon: The Canadian Phytopathological Society.
- Bayraktar H, Dolar FS. 2011. Molecular identification and genetic diversity of Fusarium species associated with onion fields in Turkey. J Phytopathol. 159:28–34.
- Chang KF, Hwang SF, Conner RL, Ahmed HU, Zhou Q, Turnbull GD, Strelkov SE, McLaren DL, Gossen BD. 2015. First report of Fusarium proliferatum causing root rotin soybean (Glycine max L.) in Canada. Crop Prot. 67:52–58.
- Conner RL, Hwang SF, Stevens RR. 1996. Fusarium proliferatum: a new causal agent of black point in wheat. Can J Plant Pathol. 18:419–423.
- Consortium TTG. 2012. The tomato genome sequence provides insights into fleshly fruit evolution. Nature. 485:635–641.
- Eken C, Demirci E, Dane E. 2004. Species of Fusarium on sainfoin in Erzurum, Turkey. New Zeal J Agr Res. 47:261–263.
- Geiser DM, Jiménez-Gasco MM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 110:473–479.
- Imazaki I, Kadota I. 2015. Molecular phylogeny and diversity of Fusarium endophytes isolated from tomato stems. FEMS Microbiol Ecol. 91:fiv098.
- Jeon YA, Yu SH, Lee YY, Park HJ, Lee S, Sung JS, KimY G, Lee HS. 2013. Incidence, molecular characteristics and pathogenicity of Gibberell afujikuroi species complex associated with rice seeds from Asian countries. Mycobiology. 41:225–233.
- Lee HB, Kim CJ, Mun HY, Choi HS, Lee YH, Yun HO. 2011. First report of crown rot on gypsophila (Gypsophila paniculata) caused by Fusarium proliferatum in Korea. Plant Dis. 95:220.
- Lee SB, Taylor JW. 1990. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide tomethods and applications. New York (NY): Academic Press; p. 282–287.
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Ames, IW: Blackwell Publishing Professional.
- McGovern RJ. 2015. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 73:78–92.
- Nelson PE, Toussoun TA, Marasas WFO. 1983. Fusarium species: an illustrated manual for identification. University Park: Pennsylvania State University Press.
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 7:103–116.
- O’Donnell K, Cigelnik E, Nirenberg HI. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 90:465–493.
- Price WD, Lovell RA, McChesney DG. 1993. Naturally occurring toxins in feedstuffs: center for veterinary medicine perspective. J Anim Sci. 71:2556–2562.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tonti S, Pra MD, Nipoti P, Prodi A, Alberti I. 2012. First report of Fusarium proliferatum causing rot of stored garlic bulbs (Allium sativum L.) in Italy. J Phytopathol. 160:761–763.
- Zhang JX, Wu XX, Bi YQ, Wu YX, Lin GH, He YQ, Mao ZC. 2013. First report of Fusarium proliferatum infecting carnation (Dianthus caryophyllus L.) in China. J Phytopathol. 161:850–854.
- Zhang M, Wang Y, Wen CY, Wu HY. 2012. First report of Fusarium proliferatum causing fruit rot of winter jujube (Zizyphus jujuba) in storage in China. Plant Dis. 96:913.
