Abstract
Pepper mottle virus (PepMoV) is a widespread threat to vegetable crop production in the USA and south-east Asia. We describe the development of a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay to detect PepMoV. The RT-LAMP assay was based on a set of four primers that match a specific region of the coat protein gene in the PepMoV genome. The detection limit of conventional RT-PCR detection was 1.47 × 10−4 µg µL−1 of cDNA, whereas RT-LAMP was 10 times more sensitive. Using RT-LAMP, PepMoV detection was also highly specific, showing no cross-activity with four other potyviruses. Sixty-nine field samples collected from symptomatic pepper plants growing in five South China provinces were tested for the presence of PepMoV infection by performing an RT-LAMP assay as well as a conventional RT-PCR. Both methods detected PepMoV in 18 samples, demonstrating that the PepMoV-specific RT-LAMP assay could be a useful alternative tool for the diagnosis and epidemiological surveillance of PepMoV infections. The RT-LAMP assay also has the advantages that it can be performed in a low-tech environment and is quicker and cheaper to perform than conventional RT-PCR.
Résumé
Le virus de la marbrure du piment (PepMoV) est une menace répandue qui touche les cultures légumières aux États-Unis et en Asie du Sud-Est. Nous décrivons la mise au point d’une épreuve RT-LAMP (test moléculaire d’amplification isothermale en temps réel) afin de détecter le PepMoV. L’épreuve a été basée sur un jeu de quatre amorces qui correspondent à une région précise du gène de la protéine capsidique dans le génome du PepMoV. La limite de détection d’une RT-PCR classique était de 1.47 × 10−4 µg µL−1 d’ADNc, tandis que l’épreuve LAMP était 10 fois plus sensible. La détection du PepMoV à l’aide de l’épreuve RT-LAMP était également très précise, n’affichant aucune activité croisée avec quatre autres potyvirus. Soixante-neuf échantillons collectés en champ sur des piments symptomatiques cultivés dans cinq provinces du sud de la Chine ont été testés pour y détecter l’infection au PepMoV à l’aide de l’épreuve RT-LAMP de même que par RT-PCR classique. Les deux méthodes ont permis de détecter le PepMoV dans 18 échantillons, démontrant que l’épreuve RT-LAMP propre au PepMoV pourrait être un outil alternatif utile pour procéder au diagnostic et à la surveillance épidémiologique des infections causées par ce virus. L’épreuve RT-LAMP offre aussi l’avantage qu’elle peut être utilisée dans un environnement technologique rudimentaire et qu’elle est plus rapide et moins chère que la RT-PCR.
Introduction
Pepper mottle virus (PepMoV) is a member of the genus Potyvirus, the largest genus of plant viruses (Shukla et al. Citation1994). PepMoV was first recognized in Arizona, USA, in 1969 (Nelson & Wheeler Citation1972) as a new strain of Potyvirus that infected peppers. PepMoV has the typical characteristics of a Potyvirus: a genome consisting of a single strand of positive-sense RNA of ~10 kb, surrounded by a capsid comprised of a single viral-encoded protein (Vance et al. Citation1992). Since its initial isolation, the occurrence of PepMoV has also been reported in other regions of the USA (Quiñones et al. Citation2011) and in several countries and regions of south-east Asia (Cheng et al. Citation2011). There is an urgent need to develop a useful and simple diagnostic tool for the surveillance of PepMoV epidemics.
RT-PCR and serology can be used for the early and rapid detection method of PepMoV infections (Abdalla & Dodds Citation1985; Lee et al. Citation2011). Although these technologies are both sensitive and highly specific, they are not readily transferable to a low-technology laboratory. Recently, a loop-mediated isothermal amplification (LAMP) assay has been used to detect plant viruses (Fukuta et al. Citation2003; Lenarčič et al. Citation2013; Okuda et al. Citation2015). This method involves the use of a specially designed primer set that recognizes at least four independent regions of the target gene to increase the specificity and rapidity of the reaction. Furthermore, the detection of viral cDNA can be visualized as a colour change or as an increase in turbidity that can be seen with the naked eye (Qiao et al. Citation2007). LAMP reactions take place under isothermal conditions and, therefore, the reaction can be performed using a simple water bath. As a result, LAMP assays are being increasingly used to detect various pathogens (Mori et al. Citation2001; Qiao et al. Citation2007; Lenarčič et al. Citation2013).
In this study, we describe an RT-LAMP procedure that specifically detects PepMoV and has the merit of being simple, efficient, cost-effective and convenient. We also demonstrate its usefulness as an alternative tool for diagnosing the presence of PepMoV in field samples.
Materials and methods
Virus sources
A PepMoV isolate (accession no. LN832375) was isolated in Hunan province of P. R. China on 13 August 2013 from a pepper plant that was showing typical mottling and wilt symptoms. Four other common potyviruses that infect pepper were also used in this study: Potato virus Y (PVY), Chilli ring spot virus (CRSV), Chilli veinal mottle virus (ChiVMV) and Pepper veinal mottle virus (PVMV). These viruses were propagated for use in the study by gently rubbing the surface of leaves of pepper plants with a preparation of the virus. The infected plants were maintained under greenhouse conditions until needed.
RNA isolation and reverse transcription
Total RNA was extracted from plant leaf tissues using TRIzol-A+ Reagents (Tiangen Biotechnol, Beijing, China) according to the manufacturer’s protocol with some minor modifications. Briefly, 100 mg of each tissue sample was homogenized in 1 mL of TRIzol reagent using liquid nitrogen. The tissue was then placed in 1.5 mL microfuge tubes and centrifuged to remove the cell debris and insoluble materials. The supernatant was purified using chloroform and the total RNA was precipitated using isopropyl alcohol before being dissolved in RNase-free water and then stored in a −80°C freezer until it was needed.
PepMoV cDNA was synthesized from the extracted total RNA using TransScript All-in-one First-Strand cDNA Synthesis SuperMix for PCR (TransGen, Beijing, China) according to the kit guide. The reaction mixture (20 μL) comprised 4 μL 5×TransScript All-in-One Super Mix for PCR, 2 μL total RNA as a template, and 14 μL RNase-free water. The reverse transcription reaction was performed by first incubating the reaction mixture at 25°C for 10 min, then 42°C for 30 min, and finally 85°C for 5 min to inactivate the reverse transcriptase. The cDNA synthesized was stored in a −20°C freezer until needed.
Optimal LAMP conditions
Several sequences of the PepMoV, PVY, CRSV, ChiVMV and PVMV coat proteins were aligned by MegAlign (data not shown). A 199 bp conserved region (nt 4180–nt 4378) (European Nucleotide Archive accession no. LN832375) of the PepMoV coat protein with relatively low homology was then selected as the template for the LAMP primers, which were designed using an online software program (http://primerexplorer.jp/elamp3.0.0/index.html; Eiken Chemical Co., Ltd, Tokyo, Japan). Primers were synthesized and purified using HPLC by HUADA Co, Ltd (Shenzhen, China).
To optimize the LAMP assay, different amplification temperatures (60–66°C) and reaction times (30–120 min) were tested using cDNA synthesized using the Loopamp© DNA Amplification Kit (Eiken China, Beijing, China) following the manufacturer’s instructions. Following optimization, the LAMP reaction was carried out in a 25 μL volume comprising 12.5 μL of 2×Reaction Mix, 1 μL of each inner primer (10 μM), 0.5 μL of each outer primer (10 μM), 1 μL of Bst DNA polymerase, and 8.5 μL of distilled water. The amplification was performed by incubating the mixture at 62°C for 75 min, followed by incubation at 80°C for 10 min to inactivate the Bst DNA polymerase.
Following amplification, 2 μL of the LAMP products were mixed with SYBR Green I and separated by performing electrophoresis in a 2% agarose gel for 30 min at a constant 100 V. Fluorexon (Mori et al. Citation2001) was used as an alternative visualization agent to enable the results to be checked by the naked eye because we and others (Goto et al. Citation2009) have observed that SYBR Green I inhibits LAMP reactions. 1.5 µL of 1:250 diluted Fluorexon was added to the reaction tubes. The presence of PepMoV cDNA was indicated by a change in colour from orange to green.
LAMP sensitivity and specificity
The concentration of the synthesized cDNA was evaluated using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). The sensitivity of the LAMP assay was tested by performing serial 10-fold dilutions of the synthesized PepMoV cDNA (1.47 × 10−1–1.47 × 10−9 μg μL−1). The specificity of the LAMP assay was assessed using the synthesized cDNA of four potyviruses: PVY, CRSV, ChiVMV and PVMV. For each experimental set, a LAMP assay with a parallel negative control lacking a template was performed.
Evaluation of the ability of the LAMP assay to detect PepMoV in field samples
In August 2014 and August 2016, the sensitivity of conventional RT-PCR (Lee et al. Citation2011) and the RT-LAMP assay were compared using field samples from different regions of South China. The field sample amplification products were evaluated using conventional PCR and sequenced by the Beijing Genomics Institute (BGI, Shenzhen, China).
Results and discussion
Development and optimization of RT-LAMP
The selected LAMP primer set (sequences shown in ) comprised four primers: two outer primers (F3 and B3), a forward inner primer FIP and a backward inner primer BIP. To maximize DNA production using RT-LAMP, the reaction conditions, including the incubation temperature and duration, needed to be optimized. As shown in , the DNA products amplified at temperatures from 60°C to 64°C at duration of 60 min had a characteristic ladder-like appearance on 2% agarose gel. The ladder-like DNA fragments were brightest at 63°C, indicating that 63°C was the optimum temperature for amplifying DNA.
Table 1. Primer set used for RT-LAMP in this study.
Fig. 1 Optimization of the reaction conditions used for RT-LAMP to detect PepMoV on an agarose gel. (a) Incubation temperatures: 60ºC, 61ºC, 62ºC, 63ºC, 64ºC, 65ºC and 66ºC. (b) Incubation duration: 30 min, 45 min, 60 min, 75 min, 90 min and 120 min. M indicates the 5 kb DNA ladder (Vazyme). NTC indicates the non-template control.
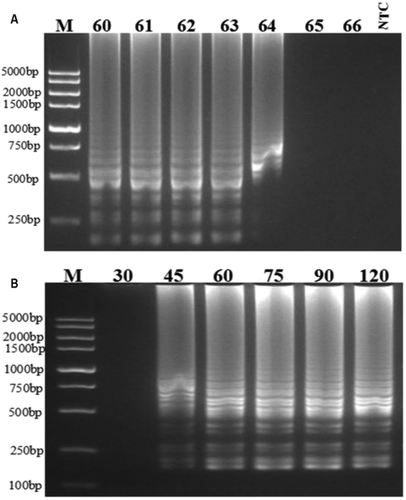
To determine the minimum time required for the RT-LAMP reaction, which was visualized by staining on an agarose gel after electrophoresis, six incubation time points were evaluated using 63°C as the incubation temperature. Amplified ladder-like DNA products were observed in agarose gel for all incubation time points except for 30 min. The brightness of the ladder-like DNA fragments gradually increased as the incubation times increased from 45 min to 75 min. Therefore, incubation at 63°C for 75 min was considered to be the optimum reaction conditions for RT-LAMP using the primers designed for this RT-LAMP assay.
Specificity assessment
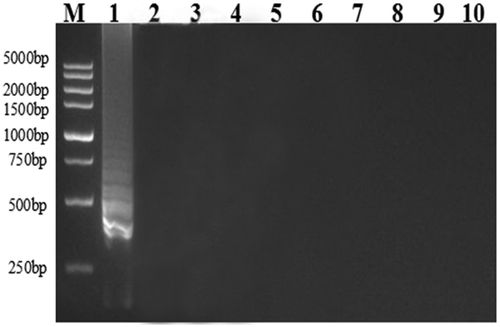
Co-infection of pepper and other Solanaceae crops by several potyviruses is a common phenomenon (Garcêz et al. Citation2015) and, therefore, it is crucial to develop an RT-LAMP assay that is specific for a particular virus. The reliability of our RT-LAMP assay depends on the specificity of the primer sets that were used. To determine the specificity of the RT-LAMP assay for PepMoV, we examined whether there was any potential cross-activity with the closely related potyviruses PVY, CRSV, ChiVMV and PVMV. Although ladder-like DNA fragments in agarose gels were observed in reactions containing PepMoV cDNA, no visible DNA product was generated in reactions containing cDNA preparations of PVY, CRSV, ChiVMV or PVMV (), indicating that the RT-LAMP assay developed for PepMoV detection was highly reliable.
Sensitivity assessment
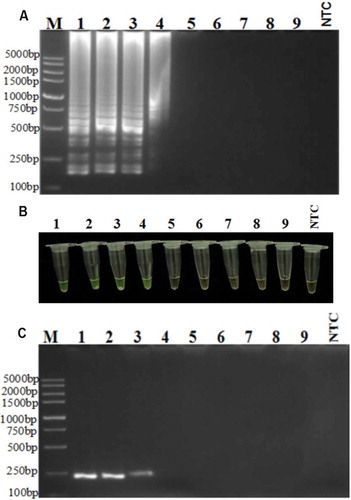
The usefulness of the RT-LAMP reaction also depends on its sensitivity (Notomi et al. Citation2000). As shown in , the LAMP assay detected PepMoV cDNA in the reaction mixture of infected pepper that had been diluted by up to 10−3, corresponding to 1.47 × 10−4 µg µL−1, whereas the conventional RT-PCR detected PepMoV cDNA in reaction mixtures that had been diluted by up to 10−2, corresponding to 1.47 × 10−3 µg µL−1, which means that the sensitivity of the LAMP assay () was 10 times greater than that of conventional RT-PCR (). This result agrees with the findings reported by previous authors (Notomi et al. Citation2000; Goto et al. Citation2009) and underlines another advantage of developing RT-LAMP assays. To simplify the detection process, positive reactions were also visualized directly by adding Fluorexon to the reaction tubes to generate a green colour, whereas the negative controls remained orange ().
Fig. 3 Sensitivity of RT-LAMP assay for PepMoV detection. (a) RT-LAMP, (b) direct staining with Fluorexon in the reaction tubes, (c) RT-PCR, using serial (10-fold) dilutions of cDNA (Lane 1, 1.47 × 10−1 µg µL−1; Lane 2, 1.47 × 10−2 µg µL−1; Lane 3, 1.47 × 10−3 µg µL−1; Lane 4, 1.47 × 10−4 µg µL−1; Lane 5, 1.47 × 10−5 µg µL−1; Lane 6, 1.47 × 10−6 µg µL−1; Lane 7, 1.47 × 10−7 µg µL−1; Lane 8, 1.47 × 10−8 µg µL−1; Lane 9, 1.47 × 10−9 µg µL−1; NTC, non-template control).
Evaluation of LAMP using suspected field samples
To verify the potential utility of a standardized RT-LAMP assay as a rapid diagnostic tool, 69 field samples were collected from pepper plants growing in five different provinces in South China that showed symptoms of mottle or mosaic on the leaves and severe stunting. The samples were tested for the presence of a PepMoV infection by performing an RT-LAMP assay and also a conventional RT-PCR. Both methods detected PepMoV in 18 of the samples (), confirming that the RT-LAMP assay could be a useful alternative diagnostic tool for PepMoV surveillance. This result also demonstrated that the PepMoV had infected pepper crops in all five of the provinces that were sampled in South China.
Table 2. Detection of PepMoV in field samples using RT-LAMP and RT-PCR.
Additional information
Funding
References
- Abdalla A, Dodds JA. 1985. Serological detection of pepper mottle virus in California pepper fields. Plant Dis. 70:173.
- Cheng YH, Deng TC, Chen CC, Liao JY, Chang CA, Chiang CH. 2011. First report of Pepper mottle virus in bell pepper in Taiwan. Plant Dis. 95:617.
- Fukuta S, Kato S, Yoshida K, Mizukami Y, Ishida A, Ueda J, Kanbe M, Ishimoto Y. 2003. Detection of tomato yellow leaf curl virus by loop-mediated isothermal amplification reaction. J Virol Methods. 112:35–40.
- Garcêz RM, Chaves ALR, Eiras M, Meletti LMM, de Azevedo Filho JA, da Silva LA, Colariccio A. 2015. Survey of aphid population in a yellow passion fruit crop and its relationship on the spread Cowpea aphid-borne mosaic virus in a subtropical region of Brazil. SpringerPlus. 4:537.
- Goto M, Honda E, Ogura A, Nomoto A, Hanaki K. 2009. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 46:167–172.
- Lee MY, Song YS, Ryu KH. 2011. Development of infectious transcripts from full-length and GFP-tagged cDNA clones of Pepper mottle virus and stable systemic expression of GFP in tobacco and pepper. Virus Res. 155:487–494.
- Lenarčič R, Morisset D, Mehle N, Ravnikar M. 2013. Fast real-time detection of potato spindle tuber viroid by RT-LAMP. Plant Pathol. 62:1147–1156.
- Mori Y, Nagamine K, Tomita N, Notomi T. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Comm. 289:150–154.
- Nelson MR, Wheeler RE. 1972. A new virus disease of pepper in Arizona. Plant Dis Rept. 56:731.
- Notomi T, Okayama H, Masubuchi H. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63.
- Okuda M, Okuda S, Iwai H. 2015. Detection of Cucurbit chlorotic yellows virus from Bemisia tabaci captured on sticky traps using reverse transcription loop-mediated isothermal amplification (RT-LAMP) and simple template preparation. J Virol Methods. 221:9–14.
- Qiao YM, Guo YC, Zhang XE, Zhou YF, Zhang ZP, Wei HP, Yang RF, Wang DB. 2007. Loop-mediated isothermal amplification for rapid detection of Bacillus anthracis spores. Biotechnol Lett. 29:1939–1946.
- Quiñones M, Arana F, Alfenas-Zerbini P, Soto M, Ribeiro D, Diaz A, Gonzalez D, Carbonell J, Depestre T, Zerbini FM. 2011. First report of Pepper mottle virus in sweet pepper in Cuba. New Dis Rep. 24:2044–2058.
- Shukla DD, Ward CW, Brunt AA. 1994. The Potyviridae. Cambridge (UK): CAB International.
- Vance VB, Moore D, Turpen TH, Bracker A, Hollowell VC. 1992. The complete nucleotide sequence of pepper mottle virus genomic RNA: comparison of the encoded polyprotein with those of other sequenced potyviruses. Virology. 191:19–30.