Abstract
Since 2005, foliar symptoms of a presumptive virus disease, including chlorotic and necrotic ring patterns or blotches, have been observed in peach (Prunus persica L.) trees in Aguascalientes, Mexico. Infected leaf samples were analysed based on general scanning for Potyvirus by dot blot and by RT-PCR for specific detection. The present study describes the identification of Plum pox virus (PPV) as the causal agent of the virus disease on peach orchards in Aguascalientes, Mexico. RFLP analyses showed that PPV Mexican strain belongs to genotype D, previously detected in North America (USA and Canada). To our knowledge, this is the first report of the presence of PPV in Mexico and this data can positively contribute to establish appropriate phytosanitary control measures, in Mexico as well worldwide.
Résumé
Depuis 2005, des symptômes foliaires d’une présumée maladie à virus, y compris des anneaux concentriques chlorotiques et nécrotiques ou des taches, ont été observés sur les pêchers (Prunus persica L.) à Aguascalientes, au Mexique. Des échantillons de feuilles infectées ont été analysés en se basant sur la détection des potyvirus par hybridation sur tache, et sur la RT-PCR pour une détection précise. Cette étude décrit l’identification du virus de la sharka en tant qu’agent causal de la maladie à virus sévissant dans les vergers de pêchers à Aguascalientes, au Mexique. Les analyses RFLP ont montré que la souche du virus mexicain de la sharka appartient au génotype D, détecté auparavant en Amérique du Nord (aux États-Unis et au Canada). À notre connaissance, il s’agit de la première mention du virus de la sharka au Mexique; et ces données peuvent vraiment contribuer à mettre au point des mesures phytosanitaires adéquates de lutte, et ce, tant au Mexique que partout ailleurs dans le monde.
Introduction
Peach (Prunus persica L., Family Rosaceae) is one of the main temperate fruit crops grown worldwide with a production of 20 million metric tons (USDA Citation2016). In Mexico, the peach cultivation area is approximately 37,000 ha with a production of up to 173,000 tons, and a market value of approximately US$70 million. The most important production areas are Chihuahua, Estado de Mexico, Michoacán, Zacatecas, Puebla, Morelos, Tlaxcala, Jalisco and Aguascalientes. The latter has a production up of 4,000 tons (SAGARPA Citation2014). Plum pox virus (PPV), the causal agent of sharka disease, is one of the most destructive diseases of stone fruits (García et al. Citation2014). About 100 million stone fruit trees in Europe are currently infected by this disease, resulting in losses of around 80% and in some cases total losses (Rimbaud et al. Citation2015). In Mexico, the presence of PPV has not been reported previously but it is registered as a potential quarantine pest (Mora-Aguilera et al. Citation1999), through regulating the importation of propagative plant material from USA and Canada, where the PPV has already been detected in infected plant material (Levy et al. Citation2000; Thompson et al. Citation2001). In this study, we confirmed the presence of PPV in peach orchards in Mexico by immunoassays and molecular techniques. The results contribute to awareness of the disease and hence the implementation of appropriate phytosanitary control measures in Aguascalientes and other parts of Mexico, as well worldwide.
Materials and methods
Sampling
Two hundred and sixty-two leaf samples were collected from five orchards located in the state of Aguascalientes, Mexico, in the centre of the country. The symptoms observed were chlorosis on leaves, ring-shaped necrotic spots, malformations of leaves and eventually dwarfism (). No symptoms on fruits were observed. The sampling was divided into two periods: first from October 2005 to July 2006 and the second from August 2006 to March 2007.
Dot blot immunoassays
Immunoassay analyses were performed by specific antibodies and processed according to the manufacturer’s instructions (Agdia, Inc., Elkhart, IN, USA). Leaf samples (150–200 mg) were frozen at −70°C and macerated in PBS buffer and centrifuged at 14,000 × g for 4 minutes at room temperature. Samples (30 μL) were fixed and neutralized in nitrocellulose paper with Potyvirus-specific antibodies (anti-Potyvirus Agdia Testing Services, Inc.) and processed in a dot blot equipment (Invitrogen Co., USA). The Papaya ringspot virus (PRSV) was used as the positive control for Potyvirus in all immunoassays (kindly donated by Dra. Laura Silva Rosales, CINVESTAV-Irapuato).
Molecular identification
RT-PCR technique was used for PPV detection and genotyping. Total RNA extraction was performed with Trizol (Invitrogen Co., USA) and the RT reaction was carried out using the protocol for Super ScriptTMFirst-Strand (Invitrogen) according to the manufacturer’s specifications. The PCR was performed according to Gibbs & Mackenzie (Citation1997). One cycle of 94°C for 2 min, 35 cycles of 94°C for one min, 35 cycles at 60°C for 2 min, 35 cycles of 72°C for 3 min, and finally one cycle of 72°C for 8 min. Amplified products were separated on a 1.5% agarose gel. Degenerate primer pairs (forward: 5´-CACGGATCCCGGG(T)17(A/C/G)GC-3 and reverse: 5´-ACCACAGGATCCGGB(G/T/C)AA(T/C)AA(T/C)AG(T/C)GG(A/G/T)CA(G/A)CC-3´ were used to amplify a fragment from the 3ʹ end of the genome. Images of the gels were captured using Chemi Doc, image analyser and the software Quantity One (Bio-Rad, CA, USA). Positive PCR amplicons were purified by spin columns using the QIAquick Gel Extraction System (Qiagen, Germany) and then subjected to nucleic acid sequencing as reported in the results section.
Genotype determination
To determine the genotype (strain) of the PPV, the methodology of RFLP described by Zagrai et al. (Citation2008) was used. Using PCR, the C-terminus of PPV capsid protein (CP) gene was amplified using primer pairs P1 (5´-CCCTCACATCACCAGAGCCA-3´) and P2 (5´-CAGACTACAGCCTCGCCAGA-3´). To differentiate PPV strains, the amplified PCR products were specifically digested with RsaI and AluI. PPV-D and PPV-M strains can be discriminated by RsaI based on the polymorphism (absence or presence of the restriction site) located in the CP region.
Pathogenicity test
To determine the pathogenic properties of the virus obtained from positive samples, a pathogenicity test was developed. Thirty plants of Nicotiana benthamiana, 12 plants of Chenopodium amaranthicolor (goosefoot) and 12 plants of Sonchus oleraceus L. (sowthistle) were used. Plants were grown under greenhouse conditions and mechanically inoculated with crude extracts from infected leaves. The inoculum was prepared by maceration with approximately 100 mg of infected leaves in 1 mL of 10 mM phosphate buffer (pH 7) and carried out in healthy plants using carborundum as the abrasive agent. Infection in the inoculated plants was evaluated by the immunoprinting leaf tissue method. For this, leaves with viral symptoms (and leaves from non-infected control plants) were cut and placed between two membranes of nitrocellulose previously neutralized with PBS buffer. Then, a force of 80 kg cm−2 was applied for 2 min. The membranes were processed using the dot blot technique as described above.
Results and discussion
Molecular identification
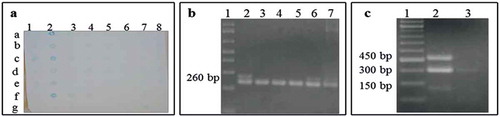
To evaluate the infection in peach samples ( and ), the viral protein was identified by dot blot technique (, ). This immunoassay analysis demonstrated the presence of Potyvirus protein in symptomatic plants in 9.5% of the samples (262) analysed. Positive PPV samples were confirmed by the amplification of a ~ 260 by RT-PCR (). Five positive PCR amplicons were sequenced and compared by BLAST with NCBI-database and corresponded with 98% nucleotide identity with PPV.
Table 1. Number of peach leaf samples collected, and those positive for Plum pox virus by dot blot or RT-PCR.
Fig. 2 (Colour online) (a) Dot blot analysis performed on samples collected from different orchards for Potyvirus scrutiny. 1/a: negative (healthy plants), 2/a: positive for Potyvirus (Papaya ringspot virus), 2/b to 8/g: samples from apparently infected peach trees. (b) RT-PCR products obtained with consensus oligos for Potyvirus. In all cases a band of 260 bp was obtained. Lanes: (1) DNA ladder, (2) positive control, and (3–7) samples of infected peach trees. (c) RsaI restriction profile in the CP region representative of PPV-D genotype by RFLP analyses. Lanes: (1) DNA ladder, (2) PPV genotype D, showing the amplicon of ~450 bp and two fragments of ~300 and ~150 bp, (3) complete digestion with RsaI.
Genotype determination
RFLP analysis showed amplified fragments of ~450 bp, and two fragments of ~300 and ~150 bp sizes (Fig. 2c), indicating that isolates correspond to PPV-genotype D. PPV-D is commonly distributed in the Americas. In North America, it was detected a decade and a half ago (Levy et al. Citation2000; Thompson et al. Citation2001) and was recently detected in South America (Rezende et al. Citation2015). There are no additional reports about the presence of PPV strains in Mexico.
Pathogenicity test
The pathogenicity test included C. amaranthicolor, S. oleraceus and N. benthamiana. After 12–14 days post-inoculation (dpi), symptoms of necrotic spots were observed on S. oleraceus and N. benthamiana (, ). Symptoms in C. amaranthicolor were not observed. Previously, yellow spots with necrotic centres were reported on Chenopodium foetidum following experimental inoculation (Glasa et al. Citation1997). Furthermore, hypersensitivity reaction has been reported in PPV experimental infections with N. benthamiana, N. clevelandii and Pisum sativum L. (Kegler et al. Citation2001).
Table 2. Pathogenicity test on three plant species using Plum pox virus inoculum and percentage of inoculation success.
In conclusion, we have identified for the first time the presence of Plum pox virus (PPV) genotype D in Mexican peach orchards in Aguascalientes, Mexico. However, further investigation is necessary to determine the possible origin of PPV in peach crops in Mexico, as well as the distribution of PPV strains in other areas in the country and possible impact on production of peaches.
Acknowledgements
This project was supported by CONACYT, from Mexico under grant N° 546667. We thank Dr Laura Silva Rosales (CINVESTAV-Irapuato) for the material kindly donated and used as the control, Dr Rodolfo Velásquez (INIFAP-Aguascalientes) for the assistance provided during sampling in the field, and Dra. Andrea Murillo Gallo for providing final editorial assistance.
Additional information
Funding
References
- García JA, Glasa M, Cambra M, Candresse T. 2014. Plum pox virus and sharka: a model potyvirus and a major disease. Mol Plant Pathol. 15:226–241.
- Gibbs A, Mackenzie A. 1997. A primer pair for amplifying of the genome of all potyvirids by RT-PCR. J Virol Meth. 63:9–16.
- Glasa M, Matisova J, Kudela O. 1997. Susceptibility of peach GF 305 seedlings and selected herbaceous plants to Plum pox virus isolates from western Slovakia. Acta Virol. 41:341–344.
- Kegler H, Grüntzig M, Fuchs E, Ranković M, Ehrig F. 2001. Hypersensitivity of plum genotypes to Plum pox virus. J Phytopathol. 149:213–218.
- Levy L, Damsteegt V, Welliver R. 2000. First report of Plum pox virus (Sharka disease) in Prunus persicae in the United States. Plant Dis. 84:202.
- Mora-Aguilera G, Levy L, Téliz D, Martínez-Gómez P, Dicenta F, Nieto-Angel R, Gutiérrez-Espinosa A. 1999. Plum pox virus: a potential quarantine pest of Mexico. Rev Chapingo Ser Hort. 5:51–58.
- Rezende JAM, Camelo-Garcia VM, Kitajima EW. 2015. First report on detection of Plum pox virus in imported peach fruits in Brazil. Plant Dis. 100:869.
- Rimbaud L, Dallot S, Gottwald T, Decroocq V, Jacquot E, Soubeyrand S, Thébuad G. 2015. Sharka epidemiology and worldwide management strategies: learning lessons to optimize disease control in perennial plants. Annu Rev Pythopathol. 53:357–378.
- SAGARPA: Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación. 2014. Servicio de Información Agroalimentaria y Pesquería (SIAP), México. Available from: http://www.gob.mx/siap
- Thompson D, McCann M, MacLeod M, Lye D, Green M, James D. 2001. First report of Plum pox potyvirus in Ontario, Canada. Plant Dis. 85:97.
- USDA: United States Department of Agriculture. 2016. Fresh Peaches and Cherries: World Markets and Trade. Available from: https://apps.fas.usda.gov/psdonline/circulars/StoneFruit.pdf
- Zagrai L, Zagrai I, Ferencz B, Gaboreanu I, Kovacs K, Petricele I, Popescu O, Pamfil D, Capote N. 2008. Serological and molecular typing of Plum pox virus isolates in North of Romania. J Plant Pathol. 90:41–46.


