Abstract
Pepper vein yellows virus (PeVYV) is a member of Polerovirus and infects solanaceous crops worldwide. In this study, a highly efficient and easy-to-use single-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for PeVYV detection. This assay used two outer (F3 and B3), two inner (FIP and BIP) and two loop (LF and LB) primers in a single reaction. The optimized reaction was determined to be at 62°C for 45 min and the whole reaction took less than 1 h. This assay was about 10 times more sensitive than the conventional one-step RT-PCR. This RT-LAMP assay should allow specific detection of PeVYV in plant tissues. We recommend this PeVYV-specific RT-LAMP assay for large scale field studies of PeVYV infection.
Résumé
Le virus à veines jaunes du piment (VVJPi) est un polérovirus qui infecte les solanacées à la grandeur de la planète. Dans cette étude, une épreuve très efficace, facile à utiliser et comportant une seule étape, basée sur la technique d’amplification isotherme en temps réel (RT-LAMP), a été conçue pour la détection du VVJPi. L’épreuve faisait appel à deux amorces externes (F3 et B3), deux amorces internes (FIP et BIP) et deux amorces boucles (LF et LB) pour une seule réaction. On a établi que la réaction optimisée s’est produite à 62°C, pendant 45 minutes, et que la réaction entière a duré moins d’une heure. Cette épreuve était environ 10 fois plus sensible que la RT-PCR classique en une étape. Cette épreuve RT-LAMP devrait permettre la détection précise du VVJPi dans les tissus des plantes. Nous recommandons cette épreuve RT-LAMP spécifique du VVJPi pour des études à grande échelle de l’infection causée par le VVJPi, réalisées au champ.
Introduction
Pepper vein yellows virus (PeVYV) is the member of the genus Polerovirus, in the family Luteoviridae. This virus was previously known as Pepper yellow leaf curl virus (PYLCV) or Pepper yellows virus (PYV) based on the symptoms in infected pepper plants (Capsicum annuum L.) (Dombrovsky et al. Citation2010). The virus was reported to infect multiple important solanaceous crops in Europe (Rast Citation1988; Villanueva et al. Citation2013), Asia and Africa (Yonaha et al. Citation1995; Knierim et al. Citation2013; Zhang et al. Citation2015). Like other poleroviruses, PeVYV replicates strictly in the phloem cells of its host plants and can be readily transmitted to other plants through aphid vectors (Aphis gossypii Glover) in a persistent manner (Yonaha et al. Citation1995). In recent years, PeVYV has been reported to cause severe yield losses in several economically important vegetable crops (Villanueva et al. Citation2013; Tan et al. Citation2015). Because the geographic distribution of PeVYV is unknown, it is difficult to estimate the economic losses caused by this virus. Consequently, establishment of a high throughput and cost-effective method for PeVYV detection is highly important.
PeVYV infection in host plants was previously diagnosed using conventional RT-PCR (Yonaha et al. Citation1995; Villanueva et al. Citation2013; Zhang et al. Citation2015) and/or Northern blot assays (Zhang et al. Citation2015). Although these two detection methods are sensitive and accurate, they require expensive equipment and several hours to accomplish. A loop-mediated isothermal amplification (LAMP) method was reported to be a time saving and easy-to-use method for plant virus detection (Notomi et al. Citation2000). This method has now been used to detect several plant viruses, such as Japanese yam mosaic virus, Pepino mosaic virus, Potato leafroll virus, Potato virus Y, Tomato chlorosis virus and several other tobacco-infecting viruses (Fukuta et al. Citation2003; Nie Citation2005; Almasi et al. Citation2012; Zhao et al. Citation2012; Ling et al. Citation2013; Karwitha et al. Citation2016).
To develop a rapid and easy-to-use method for PeVYV detection, we tested and optimized a RT-LAMP method. Results presented in this paper indicated that PeVYV can be reliably detected in field-collected pepper samples using this optimized method. In addition, the RT-LAMP method was shown to be more sensitive in detecting PeVYV infection than the conventional RT-PCR method. The time required for RT-LAMP was less than 1 hour. We conclude that this optimized RT-LAMP assay is a cost-effective and high throughput assay for PeVYV epidemiological studies.
Materials and methods
Virus sources and RNA isolation
Pepper plants infected with PeVYV were identified and collected from several fields in Hunan province, China, in 2013. Total RNA was extracted from PeVYV-infected leaves using TRIzol-A+ Reagents (TIANGEN, Beijing) as previously described (Karwitha et al. Citation2016). Each RNA sample was eluted in 40 μL RNase-free water and stored at −80°C until used. Concentration of each RNA sample was measured using a Nanodrop 2000 (Thermo, USA). Other poleroviruses used in this study included Potato leafroll virus (PLRV), Potato yellow virus (PYV), Beet chlorosis virus (BCV) and Tobacco vein distorting virus (TVDV) identified and isolated from field-collected pepper tissues.
Primers for RT-LAMP assay
Complete genome or coat protein open reading frame (CP ORF) sequences of PeVYV (KM229707, KP326573, AB594828, JX427541 and KU999109) were retrieved from GenBank database. After alignment of the sequences, a 200 bp highly conserved region within the CP ORF (nucleotide position 1197–2056) was selected. The RT-LAMP inner primers FIP and BIP (5ʹ-TTGCTCGACCTTCCTCCAACTCAGACGACGAAATGGAGGCA-3′ and 5ʹ-TCAGGATCTGTCACCTTCGGGCAGGCTTTGAGAACTCCACCT-3ʹ) and outer primers F3 and B3 (5ʹ-CACACGGCGAGGAAATCG-3ʹ and 5ʹ- ACGAAGCGTATGTTGACCAT-3ʹ), respectively, were identified using the online PrimerExplorer V4 software (http://primerexplorer.jp/elamp4.0.0/index.html). Two loop primers LF and LB (5ʹ- TCTCGGCTTCTTCGGTTCC-3ʹ and 5ʹ-TCAGAGAGCGTCGCGCTTTC-3ʹ) were designed using DNAMAN v5.0 software.
Establishment and optimization of RT-LAMP
The RT-LAMP assay was first conducted using an RNA amplification kit as instructed (Loopamp, Japan). Each 25 μL reaction contained 12.5 μL 2× reaction buffer [containing Tris-HCl (pH 8.8) 40 mM, KCl 20 mM, MgSO4 16 mM, (NH4)2 SO4 20 mM, Tween-20 0.2%, Betaine 1.6 M, dNTPs 2.8 mM], 1 μL enzyme mixture, 3.5 μL deionized H2O, 1.0 μL fluorescent detection reagent (calcein 10 mM, LoopampTM, Beijing), 1.0 μL 40 μM FIP primer, 1.0 μL 40 μM BIP primer, 1 μL 20 μM LF primer, 1 μL 20 μM LB primer, 0.5 μL 10 μM F3 primer, 0.5 μL 10 uM B3 primer and 2.0 μL total RNA. The reaction mixture was incubated in a 65°C water bath for 60 min followed by 5 min at 80°C to stop the reaction. The resulting samples were examined for positive or negative result by the naked eye (Tomita et al. Citation2008) or through electrophoresis in 2% agarose gels. The RT-LAMP protocol was then further optimized by incubating the RT reactions at 60, 62, 64, or 66°C for 60 min followed by the LAMP reaction for 30, 45, 60 or 75 min.
Sensitivity and specificity of RT-LAMP
Sensitivity of the RT-LAMP assay was determined using diluted total RNA samples (10° to 10–8) in each reaction. The results then were compared with that obtained using the conventional RT-PCR method described previously (Zhang et al. Citation2015). The final products were visualized in 2% agarose gels. Specificity of the RT-LAMP assay was determined using total RNA samples isolated from the PeVYV-, PLRV-, PYV-, BCV- or TVDV-infected pepper leaf tissues. Total RNA from an uninfected pepper leaf tissue was used as a control. The results were compared with that obtained through the conventional RT-PCR.
Detection of PeVYV in field-collected samples using the conventional RT-PCR or the optimized RT-LAMP method
Conventional RT-PCR and the optimized RT-LAMP assay were used to detect PeVYV in field pepper leaf samples collected in 2014 from different regions in the Hunan and Guangxi provinces. Results obtained by the two methods were then compared.
Results
Establishment of a RT-LAMP assay
To determine if the designed primers were specific for PeVYV, we performed our initial RT-LAMP assay using an RNA amplification kit as instructed by the manufacturer (data not shown). We then tried to modify the assay by direct addition of the calcein dye into the reaction mixture prior to the first incubation. Results of the assay (Fig. S1) showed that the reactions with either the positive control or the PeVYV-infected sample (tubes 1 and 3) turned green while the reactions with either the negative control or the healthy pepper sample (tubes 2 and 4) remained a light brown in colour, indicating that this assay is specific for PeVYV detection.
Optimization of the RT-LAMP assay
To optimize the RT-LAMP assay conditions, we tested various incubation temperatures and durations. Results of the tests showed that although positive RT-LAMP reaction was observed under 60°C, stronger reactions were obtained when the temperature was set at 62–66°C (). The results also indicated that under 62°C, 45 min incubation followed by 5 min incubation at 80°C resulted in a development of strong green color (). Interestingly, when the incubation time lasted for 75 min, the intensity of green colour in the tubes declined. Consequently, the optimized RT-LAMP condition was determined to be 45 min incubation at 62°C followed by 5 min incubation at 80°C to stop the reaction.
Fig. 1 (Colour online) Optimization of the reaction conditions in RT-LAMP for PeVYV detection. (a) Different incubation temperature; (b) Different incubation duration; (c) Sensitivity of RT-LAMP using serial (10-fold) dilutions of total RNA of PeVYV infected pepper; (d) Sensitivity of RT-PCR using serial (10-fold) dilutions of total RNA of PeVYV infected pepper; (e) Specificity using total RNA of four other viruses and PeVYV infected pepper; PLRV, Potato leafroll virus; PYV, Potato yellow virus; BCV, Beet chlorosis virus; TVDV, Tobacco vein distorting virus; M indicates DNA ladder; NTC indicates the non-template control.
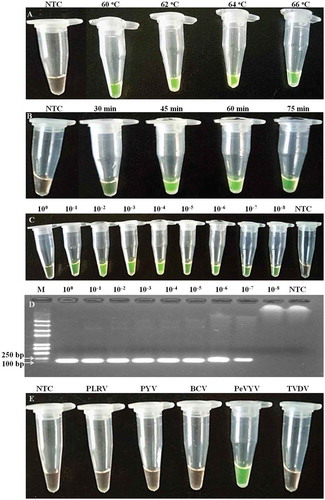
Sensitivity and specificity of the optimized RT-LAMP
Sensitivity of the optimized RT-LAMP assay was tested using diluted total RNA samples. Results of the tests showed that the optimized RT-LAMP assay could be used to detect PeVYV in the 10−8 diluted RNA samples that was about 10-fold more sensitive than that obtained by the conventional RT-PCR (Fig. 1c/d). To determine the specificity of the optimized RT-LAMP assay, we utilized total RNA samples isolated from the PeVYV-, PLRV-, PYV-, BCV- or TVDV-infected leaf tissues. Reactions without addition of RNA were used as negative controls. Results of the assays indicated that the positive reaction was only observed in tubes containing RNA samples isolated from the PeVYV-infected tissue samples (). RT-LAMP reactions using total RNA isolated from other virus-infected tissues remained a light brown in colour, similar to that shown in the negative control tube. Results obtained through the conventional RT-PCR assay agreed with the results obtained through the RT-LAMP assay, indicating that the RT-LAMP assay was indeed specific for PeVYV ().
Detection of PeVYV in field samples using the optimized RT-LAMP assay
To confirm the usefulness of this optimized RT-LAMP assay for field studies, we analysed 27 field pepper samples individually for the presence of PeVYV. The same samples were also analysed for PeVYV infection using the conventional RT-PCR assay. Results of the RT-LAMP assay showed that two of the 11 samples collected from Hunan province and nine of the 16 samples collected from Guangxi province were infected with the virus. This result was in agreement with the results obtained using the conventional RT-PCR assay (, Fig. S2 and Fig. S3). To confirm the detection result, we amplified two products from the two positive Hunan province samples using the conventional RT-PCR or the RT-LAMP, and the loop primers LF and LB. The amplified products were sequenced and analysed using BLASTn. Results showed that the two sequences shared nearly identical sequence similarity with the published PeVYV CP sequence (GenBank accession no. KP326573).
Table 1. Detection of PeVYV infection in field-collected samples using the optimized RT-LAMP or conventional RT-PCR.
Discussion
PeVYV is an important plant virus which infects many solanaceous crops in many countries (Rast Citation1988; Yonaha et al. Citation1995; Knierim et al. Citation2013; Zhang et al. Citation2015). PeVYV was first reported in red pepper in China in 2015 (Zhang et al. Citation2015) and is now known to occur in several provinces of China (Tan et al. Citation2015; Liu et al. Citation2016). To prevent the potential threat caused by PeVYV in pepper and other solanaceous crops in China, we decided to establish a detection method suitable for PeVYV epidemiological studies. In previous studies, conventional RT-PCR and Northern blot assays were employed to identify PeVYV infection in plants (Yonaha et al. Citation1995; Villanueva et al. Citation2013; Zhang et al. Citation2015). Compared with that, the LAMP was constructed for viral pathogen detection with the merits of shortening the duration, reducing the operating process as well as increasing the detection throughput (Notomi et al. Citation2000). In this paper, we report an optimized RT-LAMP assay and demonstrate that this assay can be used to detect PeVYV infection in pepper effectively and accurately. The new assay could shorten the time and manpower needed, compared with the traditional assay (Yonaha et al. Citation1995; Villanueva et al. Citation2013; Zhang et al. Citation2015), and it is convenient to operate and the results are also easily observed.
Success of a RT-LAMP assay often depends on a proper incubation temperature and amplification duration. We show in this paper that the optimal incubation temperature for this RT-LAMP assay was 62–64°C. When the reaction temperature was set at 60°C or 66°C, the intensity of reaction was weakened. This observation was in agreement with the result (63–65°C) reported for the Bst DNA polymerase (Notomi et al. Citation2000). The optimal amplification duration for the optimized RT-LAMP assay was 45–60 min after addition of the LF and LB loop primers. It is highly possible that loop primers positively impacted the speed of PeVYV detection as it was previously reported that loop primers accelerate the rate of LAMP reaction (Nagamine et al. Citation2002). Because the RT-LAMP assay can be visualized by the naked eye, this method should be suitable for laboratories lacking PCR and electrophoresis equipment and/or conducting large-scale field studies.
To confirm the specificity of the RT-LAMP for PeVYV detection, we tested samples infected with different poleroviruses. Results of the test indicated that the optimized RT-LAMP assay was indeed specific for PeVYV (). Compared with the conventional RT-PCR, this optimized RT-LAMP assay was also more sensitive (Fig. 1c/d), which was similar to previously developed LAMP assays for detecting other plant viruses (Fukuta et al. Citation2003; Karwitha et al. Citation2016). It was suggested that cross-contamination between test samples often resulted in false positive reactions during LAMP (Notomi et al. Citation2000). Karwitha et al. (Citation2016) eliminated the risk of cross contamination by keeping the reaction tubes sealed during the whole incubation process and by adding a drop of SYBR Green I to the inner cover of the reaction tube before sealing. Because direct addition of SYBR Green I into the reaction mixture might interfere with the LAMP reaction (Goto et al. Citation2009), we decided to replace SYBR Green I with calcein during our RT-LAMP assays. Bhat and co-workers (Citation2013) observed that pre-addition of calcein into the LAMP reaction mixture had no clear inhibition effect on the reaction.
In conclusion, we consider that this newly developed RT-LAMP assay is a sensitive, easy-to-use and cost-effective assay for PeVYV detection. This assay can be used by research laboratories that are engaged in epidemiological studies and without expensive equipment.
Supplemental Figure Captions
Download MS Word (14.5 KB)Supplemental Figure 3
Download JPEG Image (88.7 KB)Supplemental Figure 2
Download JPEG Image (95.6 KB)Supplemental Figure 1
Download JPEG Image (66.7 KB)Acknowledgements
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (no. 201303028), the National Natural Science Foundation of China (31571982) and the Agriculture Research System of China (CARS-25-B-05).
Supplemental material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2017.1316774
Additional information
Funding
References
- Almasi MA, Moradi A, Nasiri J, Karami S, Nasiri M. 2012. Assessment of performance ability of three diagnostic methods for detection of Potato leafroll virus (PLRV) using different visualizing systems. Appl Biochem Biotechnol. 168:770–784.
- Bhat AI, Siljo A, Deeshma KP. 2013. Rapid detection of piper yellow mottle virus and cucumber mosaic virus infecting black pepper (Piper nigrum) by loop-mediated isothermal amplification (LAMP). J Virol Methods. 193:190–196.
- Dombrovsky A, Glanz E, Pearlsman M, Lachman O, Antignus Y. 2010. Characterization of Pepper yellow leaf curl virus, a tentative new Polerovirus species. Phytoparasitica. 38:477.
- Fukuta S, Iida T, Mizukami Y, Ishida A, Ueda J, Kanbe M, Ishimoto Y. 2003. Detection of Japanese yam mosaic virus by RT-LAMP. Arch Virol. 148: 1713–1720.
- Goto M, Honda E, Ogura A, Nomoto A, Hanak KI. 2009. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. BioTechniques. 46: 167–172.
- Karwitha M, Feng ZK, Shen Y, Essendi W, Zhang WN, Li JY, Tao XR. 2016. Rapid detection of Tomato chlorosis virus from infected plant and whitefly by one-step reverse transcription loop-mediated isothermal amplification. J Phytopathol. 164:255–263.
- Knierim D, Tsai WS, Kenyon L. 2013. Analysis of sequences from field samples reveals the presence of the recently described Pepper vein yellows virus (genus Polerovirus) in six additional countries. Arch Virol. 158:1337–1341.
- Ling KS, Li R, Bledsoe M. 2013. Pepino mosaic virus genotype shift in North America and development of a loop-mediated isothermal amplification for rapid genotype identification. Virol J. 10:117.
- Liu MY, Liu XN, Li W, Zhang DY, Dai LY, Tang QJ. 2016. Complete genome sequence of a Chinese isolate of Pepper vein yellows virus and evolutionary analysis based on the CP, MP and RdRp coding regions. Arch Virol. 161:677–683.
- Nagamine K, Hase T, Notomi T. 2002. Accelerated reaction by loop mediated isothermal amplification using loop primers. Mol Cell Probes. 16:223–229.
- Nie XZ. 2005. Reverse transcription loop-mediated isothermal amplification of DNA for detection of Potato virus Y. Plant Dis. 89:605–610.
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63.
- Rast ATB. 1988. Occurence of Pepper yellow vein virus in the Netherlands. Neth J Plant Path. 94:311–313.
- Tan WP, Dong YZ, Sun XH, Liang YC, Liu HX, Zhu XP. 2015. The first identification of Pepper vein yellows virus in Shandong province, China. Plant Dis. 99:1288.
- Tomita N, Mori Y, Kanda H, Notomi T. 2008. Loop-Mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 3:877–882.
- Villanueva F, Castillo P, Isabel-Font M, Navas-Castillo J. 2013. First report of Pepper vein yellows virus infecting sweet pepper in Spain. Plant Dis. 97:1261.
- Yonaha T, Toyosato T, Kawano S, Osaki T. 1995. Pepper vein yellows virus, a novel luteovirus from bell pepper plants in Japan. Ann Phytopathol Soc Jpn. 61:178–184.
- Zhang SB, Zhao ZB, Zhang DY, Liu Y, Luo XW, Liu J, Wu LF, Peng J. 2015. First report of Pepper vein yellows virus infecting red pepper in mainland China. Plant Dis. 99:1190.
- Zhao L, Cheng J, Hao X, Tian X, Wu Y. 2012. Rapid detection of tobacco viruses by reverse transcription loop-mediated isothermal amplification. Arch Virol. 157:2291–2298.
