Abstract
The recessive Xanthomonas oryzae pv. oryzae (Xoo) resistance gene, xa25, which mediates race-specific resistance to the Xoo isolate PXO339, has previously been characterized in rice (Oryza sativa L.). Its dominant allele, Xa25, is transcriptionally induced by Xoo infection, which in turn results in susceptibility to PXO339. Utilizing a base substitution (−40, G to T) in the xa25/Xa25 promoters, a cleaved amplified polymorphic sequence (CAPS) marker was designed for effective identification of the xa25 gene. This marker co-segregated with the xa25 gene and explicitly distinguished the genotypes of xa25/Xa25 gene in rice. The marker was further tested on nine Chinese rice landraces, revealing the association of xa25/Xa25 genotypes with resistance/susceptibility to PXO339, respectively. Therefore, this marker is reliable and cost-effective for marker-assisted selection of the xa25 gene in rice.
Résumé
Le gène récessif de résistance Xanthomonas oryzae pv. oryzae (Xoo), xa25, qui médie la résistance spécifique de la race quant à l’isolat PXO339 de Xoo, a préalablement été caractérisé chez le riz (Oryza sativa L.). Son allèle dominant, Xa25, est induit transcriptionnellement par l’infection causée par Xoo qui, à son tour, engendre une sensibilité à PXO339. En utilisant une substitution de base (−40, G à T) chez les promoteurs de xa25/Xa25, un marqueur du polymorphisme de restriction des produits d’amplification (CAPS) a été conçu pour identifier efficacement le gène xa25. Ce marqueur a coségrégué avec le gène xa25, ce qui a permis de distinguer catégoriquement les génotypes du gène xa25/Xa25 chez le riz. Le marqueur a en outre été testé sur neuf variétés locales de riz chinois, ce qui a révélé le lien des génotypes xa25/Xa25 avec la résistance et la sensibilité à PXO339, respectivement. En conséquence, ce marqueur est efficace et rentable en ce qui a trait à la sélection à l’aide de marqueurs du gène xa25 chez le riz.
Introduction
Bacterial blight (BB) of rice (Oryza sativa L.), caused by Xanthomonas oryzae pv. oryzae (Xoo) (Ishiyama) (Swings et al. Citation1990), is one of the major rice diseases in the world. Using disease resistance (R) genes has been an effective and economical strategy to control BB. To date, more than 38 R genes conferring resistance to Xoo have been identified, and 26 of these genes have been mapped onto different rice chromosomes using DNA markers, and nine genes (Xa1, Xa3/Xa26, xa5, Xa10, xa13, Xa21, Xa23, xa25, Xa27) have been characterized (http://www.ricedata.cn/gene/gene_xa.htm) (Liu et al. Citation2011; Tian et al. Citation2014; Wang et al. Citation2015). Some BB R genes, such as Xa4, xa5, xa13, Xa21 and Xa23 have been pyramided to develop resistant rice cultivars using marker-assisted-selection (MAS) breeding (Lin et al. Citation1996; Suh et al. Citation2013). MAS uses genetic markers to facilitate the identification of genes of interest in a collection of diverse genotypes (Dubcovsky Citation2004). Markers targeting polymorphisms within a desired gene can reliably be utilized in the identification of desired genes in MAS (Andersen & Lübberstedt Citation2003). Based on the gene or promoter sequence of BB R gene, STS (sequence tagged site), Indel (insertion & deletion) or CAPS (cleaved amplified polymorphic sequence) markers were developed for MAS in rice BB-resistance breeding, e.g. Xa3/Xa26 (Hur et al. Citation2013), xa5 (Iyer-Pascuzzi & McCouch Citation2007), xa13 (Chu et al. Citation2006), Xa21 (Ma et al. Citation2001) or Xa27 (Yu et al. Citation2015).
The recessive R gene xa25 was identified from rice cultivar ‘Minghui63', which is an elite restorer line that has been extensively used in hybrid rice in China, and mediates race-specific resistance to the Xoo isolate PXO339 (Liu et al. Citation2011). To identify xa25/Xa25 gene in different genetic backgrounds in the transgenic rice, Liu et al. (Citation2011) used a CAPS marker for ‘Minghui63' and an Indel marker for ‘Zhonghua11' based on the sequence polymorphism in-between xa25 and Xa25 genes. However, it remains unclear that these two markers are widely used for MAS in other rice varieties. The objective of this research was to develop a DNA marker for efficient and precise identification of xa25 gene in rice.
Materials and methods
Plant materials
All rice plants were obtained from the Chinese National Rice Research Institute (Hangzhou, China) and grown in the field during June to October, 2015. At rice booting stage, the stamens of cultivar ‘IR24' were emasculated artificially by scissors and crossed with the pollen of cultivar ‘Minghui63', and their F1 plants were self-crossed to develop an F2 population.
Xoo inoculation
Xoo strain PXO339 obtained from the Chinese Academy of Agricultural Sciences (Beijing, China) was cultured on agar medium containing 20 g sucrose, 5 g peptone, 0.5 g Ca(NO3)2, 0.43 g Na2HPO4, and 0.05 g FeSO4 L−1, and grown at 28°C for 3 to 4 days. The bacterial lawn was suspended in sterile distilled water at an OD600 = 1.0 or ~109 CFU mL−1 and immediately used for plant inoculations. At the booting stage in the field (30–35°C), rice plants of ‘Minghui63', IR24 and their F2 population, and the Chinese landraces were inoculated with the PXO339 isolate using the leaf-clipping method (Kauffman et al. Citation1973). After 2 weeks of incubation in the field (30–35°C), the lesions on cut leaves were measured for identification of resistant or susceptible phenotype.
DNA extraction, PCR and restriction endonuclease digestion
Rice genomic DNA was extracted from leaves using the method described by Panaud et al. (Citation1996). The GoTaq® Master Mix (Promega, USA) was used for PCR of marker xa25-M with the primers 5ʹ-TGGCTATGGCTAGTGAGAGGT-3ʹ and 5ʹ-CTTTGTGGAGGGAAGGGAGA-3ʹ according to the manufacturer’s instructions. The PCR was performed under the following conditions: (1) 94°C for 5 min; followed by 35 cycles of (2) 94°C for 30 s, 58°C for 30 s and 72°C 30 s; and finally, (3) 72°C for 5 min. The PCR products were digested by restriction enzyme Mwo_I (New England Biolabs, Ipswich, MA) following the manufacturer’s instructions and the digests were examined via 3% agarose-gel electrophoresis and stained with ethidium bromide.
Results
Development of a CAPS marker for xa25 gene
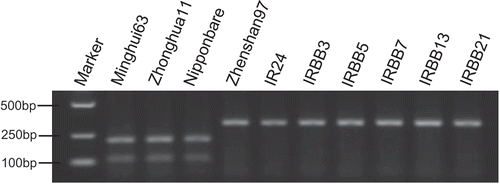
Based on the promoter sequences of xa25/Xa25 reported by Liu et al. (Citation2011), we found that a substitution (G to T) at the −40 base position leads to a different digestion pattern when using the restriction enzyme Mwo_I. Therefore, we developed a CAPS marker named xa25-M utilizing this single nucleotide polymorphism (SNP) (). To establish whether marker xa25-M can distinguish xa25/Xa25 genes, three cultivars with xa25 gene (‘Minghui63', ‘Zhonghua11' and ‘Nipponbare’) and seven cultivars without xa25 gene (‘Zhenshan97', ‘IR24', ‘IRBB3', ‘IRBB5', ‘IRBB7', ‘IRBB13' and ‘IRBB21') were tested. In the results shown in , a 344-bp band was detected in the Xa25 cultivars, whereas that of the xa25 cultivars was digested by Mwo_I into two small bands (247 bp and 103 bp). This result showed that xa25-M could clearly distinguish the xa25/Xa25 gene in these tested rice cultivars.
Fig. 1 Promoter sequences alignment of xa25 from ‘Minghui63' and Xa25 from ‘Zhenshan97'. The sequences of recognition site of MwoI are underlined. The −40 (G to T) substitution is boxed. The sequences of promoters are used by upper-case letters, and of 5ʹ UTRs are used by lower-case letters. Arrows indicate the location of the PCR primers for marker xa25-M.

Linkage analysis of the CAPS marker
To verify whether the marker xa25-M is closely linked to xa25/Xa25 gene, an F2 population with 953 plants was developed from a cross between ‘Minghui63' and ‘IR24'. F2 plants were then inoculated with Xoo isolate PXO339. Based on the lesion length invaded by Xoo, the F2 plants could be clearly classified as either resistant (< 3 cm) or susceptible (>10 cm). However, it is well known that ‘Minghui63' carries two BB R genes, xa25 and Xa3/Xa26, both against PXO339. Therefore, the resistant F2 plants were firstly analysed using the DNA marker Xa26P described by Liu et al. (Citation2011) to determine whether the plants harboured the Xa3/Xa26 gene (data not shown). Then, 168 resistant F2 plants without Xa3/Xa26 gene and 65 susceptible F2 plants were further analysed by xa25-M. As a result (), all of the resistant plants only showed the bands of xa25 genotype, and in contrast, all of the susceptible plants showed the bands of Xa25 genotype. It indicated that the genotype of xa25-M marker co-segregated with the PXO339-resistance phenotype in F2 plants. In addition, the xa25-M was a co-dominant marker which clearly differentiated heterozygous (Xa25/xa25) from homozygous (Xa25/Xa25 or xa25/xa25) plants ().
Validation of the CAPS marker by Chinese landraces
Nine Chinese rice landraces were examined to validate the marker xa25-M. Their resistant or susceptible phenotypes to PXO339 and their genotypes of marker xa25-M are shown in . Among these, four landraces were determined to be resistant to PXO339, and all of these showed a homozygous resistant genotype based on xa25-M. In contrast, the other five landraces, which were susceptible to PXO339, showed a homozygous susceptible genotype based on xa25-M.
Table 1. Phenotype of nine Chinese rice landraces genotyped with xa25-M marker.
Discussion
The xa25/Xa25 genes encode OsSWEET13, a MtN3/saliva protein, and the expression of susceptible Xa25 is induced by PXO339 (Liu et al. Citation2011). Based on the phylogenetic analysis of OsSWEET family, two susceptible genes Os11N3 and Xa13/Os8N3, encoding OsSWEET14 and OsSWEET11, respectively, are closely related to the Xa25 (Yuan & Wang Citation2013). The expression of Os11N3 is driven by the transcription activator-like (TAL) effectors AvrXa7 and PthXo3 from Xoo, which interact to a binding element in the Os11N3 promoter (Antony et al. Citation2010). Gene Xa13/Os8N3 is transcriptionally activated by another TAL effector, PthXo1, whereas this does not occur in resistant xa13 due to promoter mutation (Chu et al. Citation2006; Yang et al. Citation2006). These findings suggest that the promoter can be a key factor of the OsSWEET gene family such as Xa13/Os8N3, Os11N3 or Xa25 in mediated Xoo-rice interaction. According to the SNP in the xa25/Xa25 promoter, we designed the co-dominant marker xa25-M which was efficiently utilized to identify xa25-mediated resistance to Xoo in rice. To date, pyramiding the xa25 gene in rice cultivars has not been reported. Rice breeders could use this DNA marker to develop pyramidal lines with the xa25 gene in rice breeding.
Acknowledgements
Funding for this project was provided by the Ministry of Agriculture of China under Grant 2016ZX08009003-001; National Natural Science Foundations of China under Grant 31371593 and 31671650; and Natural Science Foundations of Zhejiang Province of China under Grant LZ14C130001 and LY13C130008.
Additional information
Funding
References
- Andersen JR, Lübberstedt T. 2003. Functional markers in plants. Trends Plant Sci. 8:554–560.
- Antony G, Zhou J, Huang S, Li T, Liu B, White F, Yang B. 2010. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 22:3864–3876.
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C. 2006. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Gene Dev. 20:1250–1255.
- Dubcovsky J. 2004. Marker-assisted selection in public breeding programs: the wheat experience. Crop Sci. 44:1895–1898.
- Hur YJ, Jeung J-U, Kim SY, Park H-S, Cho J-H, Lee JY, Sohn Y-B, Song YC, Park D-S, Lee C-W, et al. 2013. Functional markers for bacterial blight resistance gene Xa3 in rice. Mol Breed. 31:981–985.
- Iyer-Pascuzzi AS, McCouch SR. 2007. Functional markers for xa5-mediated resistance in rice (Oryza sativa, L.). Mol Breed. 19:291–296.
- Kauffman H, Reddy A, Hsieh S, Merca S. 1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas-oryzae. Plant Dis Rep. 57:537–541.
- Lin X, Zhang D, Xie Y, Gao H, Zhang Q. 1996. Identifying and mapping a new gene for bacterial blight resistance in rice based on RFLP markers. Phytopathology. 86:1156–1193.
- Liu Q, Yuan M, Zhou Y, Li X, Xiao J, Wang S. 2011. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34:1958–1969.
- Ma B, Wang W, Zou J, Liu G, Fang J, Zhu L, Zhai W. 2001. Efficient selection of homozygous lines of hybrid rice restorer with the transgene Xa21 using test cross and PCR. Chin Sci Bull. 46:125–128.
- Panaud O, Chen X, McCouch SR. 1996. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet. 252:597–607.
- Suh J-P, Jeung J-U, Noh T-H, Cho Y-C, Park S-H, Park H-S, Shin M-S, Kim C-K, Jena KK. 2013. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. 6:5.
- Swings J, Van Den Mooter M, Vauterin L, Hoste B, Gillis M, Mew TW, Kersters K. 1990. Reclassification of the causal agents of bacterial blight (Xanthomonas campestris pv. oryzae) and bacterial leaf streak (Xanthomonas campestris pv. oryzicola) of rice as pathovars of Xanthomonas orzae (ex Ishiyama 1922) sp. nov., nom. rev. Int J Syst Bacteriol. 40:309–311.
- Tian D, Wang J, Zeng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Luo Y, Murata-Hori M, et al. 2014. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 26:497–515.
- Wang C, Zhang X, Fan Y, Gao Y, Zhu Q, Zheng C, Qin T, Li Y, Che J, Zhang M, et al. 2015. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant. 8:290–302.
- Yang B, Sugio A, White FF. 2006. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA. 103:10503–10508.
- Yu P, Wang X, Yuan X, Wang C, Xu Q, Feng Y, Yu H, Wang Y, Wei X. 2015. Assessment of a functional marker for bacterial blight resistance gene locus Xa27 in rice. J Plant Pathol. 97:511–513.
- Yuan M, Wang S. 2013. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 6:665–674.