Abstract
Leaf samples from Malva parviflora plants exhibiting vein yellowing and chlorosis were collected during 2013–14 from Lahore, Pakistan and analysed for the presence of begomovirus and associated DNA satellites through PCR and subsequent sequencing. The identified full-length begomovirus genome shared maximum nucleotide (nt) sequence identity at 92.5% with Hollyhock leaf curl virus (HoLCV), representing a new strain, which was tentatively designated as HoLCV-Mal. The betasatellite shared maximum nt sequence identity (92.8%) with Kenaf leaf curl betasatellite (KLCuB) and represents a new isolate of KLCuB. The two identified alphasatellites were 86% identical to each other and shared maximum nt sequence identities at 94.3 and 93.2% with Ageratum conyzoides symptomless alphasatellite (ACSLA) and Ageratum yellow vein India alphasatellite (AYVIA), respectively. Thus, these are the new isolates of ACSLA and AYVIA identified from M. parviflora in Pakistan. To the best of our knowledge, this is the first report of a complete monopartite begomovirus complex from M. parviflora in Pakistan.
Résumé
Des échantillons de feuilles de Malva parviflora affichant de la jaunisse nervaire et de la chlorose ont été collectés en 2013 et 2014 à Lahore, au Pakistan, et analysés par PCR et, par la suite, par séquençage afin d’y détecter un bégomovirus et les ADN satellites qui lui sont associés. Le génome entier du bégomovirus partageait une identité de séquence nucléotidique maximale de 92.5% avec le virus de la jaunisse de la rose trémière (HoLCV), ce qui constitue une nouvelle souche provisoirement nommée HoLCV-Mal. Le bêta satellite partageait une identité de séquence nucléotidique maximale (92.8%) avec le bêta satellite de la jaunisse du kénaf (KLCuB), ce qui en fait donc un nouvel isolat du KLCuB. Deux alpha satellites identifiés étaient identiques à 86% et partageaient des identités de séquences nucléotidiques maximales de 94.3 et 93.2% avec l’alpha satellite asymptomatique Ageratum conyzoides (ACSLA) et l’alpha satellite indien de la rhizomanie de l’agératum (AYVIA), respectivement. Ce sont donc de nouveaux isolats d’ACSLA et d’AYVIA qui ont été identifiés à partir de M. parviflora au Pakistan. À notre connaissance, il s’agit de la première mention d’un complexe entier de bégomovirus monopartite de M. parviflora au Pakistan.
Introduction
Geminiviruses (Family Geminiviridae) are plant-infecting arthropod-transmitted viruses with circular single-stranded DNA (cssDNA) genomes (Brown et al. Citation2012). They have been classified into nine genera, namely Becurtovirus, Begomovirus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocuvirus and Turncurtovirus on the basis of host range, insect vector and genome organization (Zerbini et al. Citation2017). The members of the genus Begomovirus are the most destructive phytopathogens, which exclusively infect dicotyledonous species in the warmer parts of the world (Brown et al. Citation2015). They are transmitted by whiteflies (Bemisia tabaci) either as mono- (~2.8 kb single genomic component DNA-A) or bi-partite genomic components (two genomic components, DNA-A and DNA-B, ~2.7 kb each) (Brown et al. Citation2012; King et al. Citation2012). Most of the begomoviruses prevailing in the Old World (OW; Africa, Asia and Europe) have monopartite genomes and their DNA-A encodes six open reading frames (ORFs), two in virion-sense and four in the complementary-sense orientation, all of which are transcribed from a common bi-directional promoter in the conserved region (CR) (Rojas et al. Citation2005; Hanley-Bowdoin et al. Citation2013). However, DNA-B encodes a nuclear shuttle protein (BV1) and movement protein (BC1) in the opposite orientations (Jeske Citation2009; Hanley-Bowdoin et al. Citation2013). The majority of the OW monopartite begomoviruses are mostly found associated with betasatellite and alphasatellite molecules. The betasatellites are about half of the size of their helper begomovirus and possess cssDNA genomic structure. Betasatellites are quite flexible in their trans-replication by diverse begomoviruses; however, these molecules rely exclusively upon the helper begomovirus for their replication, movement and encapsidation (Briddon & Stanley Citation2006). Betasatellites encode a single symptom determinant protein in the complementary sense (βC1), which performs vital functions in a successful begomovirus infection. Other conserved structural features of betasatellites are an Adenine-rich (A-rich) region and a satellite conserved region (SCR) having a stem-loop (Briddon & Stanley Citation2006). The begomoviruses may or may not be able to cause infection in the plant hosts without betasatellites (Kon & Gilbertson Citation2011). These begomovirus complexes are occasionally found associated with another type of self-replicating, cssDNA satellite-like molecule known as alphasatellites (Briddon et al. Citation2004; Nawaz-ul-Rehman & Fauquet Citation2009). These molecules are ~1.4 kb in size and depend upon their helper virus for encapsidation and insect transmission, although their precise role has not been discovered yet. Most recently, another class of DNA-satellites called deltasatellite have also been reported from the New World (Fiallo-Olivé et al. Citation2016).
The role of weed hosts as an inoculum source and recombination reservoirs in begomovirus evolution is of prime importance. Marshmallow or cheese weed (Malva parviflora L., Family Malvaceae) is native to Southern Europe and Asia (Akbar et al. Citation2014) and is widespread in the major cultivated and non-cultivated agriculture ecosystems (Barros et al. Citation2010). Malva parviflora has extensive medicinal and pharmacological uses (Heinrich et al. Citation2012). Weed plants of the family Malvaceae have been serving as a reservoir in broadening the genetic diversity of many geminiviruses (Paprotka et al. Citation2010). Malva parviflora has reportedly been an alternate and/or experimental host for bipartite begomoviruses such as Abutilon mosaic Brazil virus (Paprotka et al. Citation2010), Macroptilium yellow mosaic Florida virus (Idris et al. Citation2003), South African cassava mosaic virus (Berrie et al. Citation2001) and Abutilon mosaic virus (Jeske & Werz Citation1980).
The objective of this study was to characterize the potential begomovirus infecting M. parviflora plants.
Materials and methods
Sample collection and total genomic DNA isolation
During 2013–14, leaf samples of two M. parviflora plants exhibiting typical begomovirus disease symptoms (vein yellowing and chlorosis) () were collected from the vicinity of the University of the Punjab, Lahore, Pakistan (31.29°N, 74.17°E). Total genomic DNA was isolated from the infected leaf samples using the CTAB method (Doyle & Doyle Citation1990).
Amplification, cloning and sequencing of the begomovirus complex
The viral circular DNA was enriched using rolling circle amplification (RCA) with Ф29DNA polymerase (ThermoScientific) as described previously (Haible et al. Citation2006; Qurashi et al. Citation2017). The resultant RCA products were diluted and then employed in PCR amplifications of the begomovirus complex. The initial begomoviral detection was carried out using a degenerate PCR primers pair (AC1048/AV494), to amplify ~579 bp fragment of the CP gene (Wyatt & Brown Citation1996) and further by subsequent sequencing. On the basis of the resultant CP sequences, an abutting primer pair (SonchA_F/SonchA_R; ) was designed to amplify the full-length begomovirus genome from the positive sample Sonch31. Moreover, universal primer pairs Beta01/Beta02 (Briddon et al. Citation2002) and DNA101/DNA102 (Bull et al. Citation2003) were employed to amplify alpha- and betasatellites from the same sample, respectively. Initially, Beta01/Beta02 primers were not yielding a full-length betasatellite. Subsequently, on the basis of partial sequences, an additional pair of primers (SOB_F and SOB_R; ) was designed to amplify any full-length betasatellite. All the amplicons were subsequently cloned into TA cloning vector pTZ57R/T (InsTAclone PCR cloning kit, ThermoScientific) and sequenced in their entirety from First BASE Laboratories Sdn Bhd, Malaysia.
Table 1. Primer sequences used in this study.
Sequence comparisons and phylogenetic analysis
All the obtained sequences were initially analysed using BLASTn with already submitted sequences available in the NCBI GenBank database (http:www.ncbi.nlm.nih.gov). Later, Sequence Demarcation Tool (SDT) (Muhire et al. Citation2014) was employed to determine pairwise nucleotide (nt) sequence identities with the most closely related begomovirus and DNA-satellites sequences retrieved from GenBank, respectively. The evolutionary relatedness was determined by constructing phylogenetic dendrograms in MEGA7 software (Kumar, Stecher et al. Citation2016) using Neighbour-Joining (NJ) algorithm. The coding regions were analysed in the complementary and virion sense orientations using ORF finder tool available at NCBI (http://www.ncbi.nlm.nih.gov) and are listed in with the predicted proteins.
Table 2. Predicted open reading frames (ORFs) for the begomovirus and associated DNA-satellites identified from Malva parviflora.
Recombination analysis
The begomovirus and alphasatellite sequences were further analysed for the detection of any recombination event with Recombination Detection Program (RDP4). The recombination analysis was supported by multiple algorithms available in RDP4 including BootScan (Martin et al. Citation2005), Chimera (Posada & Crandall Citation2001), GENECONV (Padidam et al. Citation1999), MaxChi (Smith Citation1992), RDP (Martin & Rybicki Citation2000) and SiScan (Gibbs et al. Citation2000). The highest acceptable P-value was set at 1 × 10−5 to predict any putative recombination breakpoint.
Results and discussion
Sequence comparison/organization and phylogenetic analysis of the full-length begomovirus genome
The full-length begomovirus clone Sonch31A, deposited in GenBank (Acc. No. LT716980) had typical OW begomovirus genome size (2755 bp) and characteristically contained all the features described for the previously reported OW monopartite begomoviruses (). The Rep of Sonch31A contained a predicted iteron-related domain (IRD) ‘FQID’ on its N-terminal, similar to Croton yellow vein virus (CrYVV) (Hussain et al. Citation2011). The IRD is believed to interact with the begomovirus genome and triggers begomovirus DNA replication. The clone Sonch31A showed maximum nt sequence identity at 92.5% with Hollyhock leaf curl virus (HoLCV) (FR772082) identified from Pakistan and infecting hollyhock (Alcea rosea). Thus, following the begomovirus species demarcation limit fixed at <91% and guidelines for their nomenclature by the International Committee for the Taxonomy of Viruses (ICTV) (Brown et al. Citation2015), the clone Sonch31A is considered a new strain of HoLCV from M. parviflora. The tentative descriptors for the new strain are suggested as Hollyhock leaf curl virus-Malva [Pakistan:Sonch31A:Malva:17] and abbreviated as HoLCV-Mal [PK:Sonch31A:Mal:17]. The new begomovirus strain HoLCV-Mal was grouped into a well-supported clade (100% bootstrap value) with HoLCV ().
Sequence comparison/organization and phylogenetic analysis of the full-length DNA-satellites
The clone Sonch31B2 was 1352 nt in length and characteristically similar to the already reported betasatellites in having an intergenic region (IR) (TAATATTA/C), a single ORF called βC1 in the complementary-sense orientation and an A-rich region. The betasatellite clone sequence shared maximum nt sequence identity at 92.8% with Kenaf leaf curl betasatellite (KLCuB; Acc. No. KT948077) from false mallow (Malvastrum coromandelianum) in Pakistan. Thus, following the suggested species demarcation threshold for betasatellite >79% (Briddon et al. Citation2008) Sonch31B2 represents a new isolate of KLCuB (Acc. No. LT716981). The phylogenetic analysis grouped Sonch31B2 into a well-supported clade (99% bootstrap support) with KLCuB ().
Fig. 2 Phylogenetic dendrograms based on complete nt sequences of (a) begomovirus, (b) betasatellite, and (c) alphasatellite isolates, using Neighbour-Joining (NJ) algorithm in MEGA7. All the isolates identified from Pakistan in this study are shown in bold white text on black background. Horizontal lines represent nt substitutions per site. Numeric values at branch nodes are representing per cent bootstrap values higher than 60 (1000 replicates). All isolates used for comparison are represented by their respective accession numbers in the trees. All abbreviations for begomovirus, alpha- and betasatellite isolates are according to Brown et al. (Citation2012) and Briddon et al. (2012), respectively.
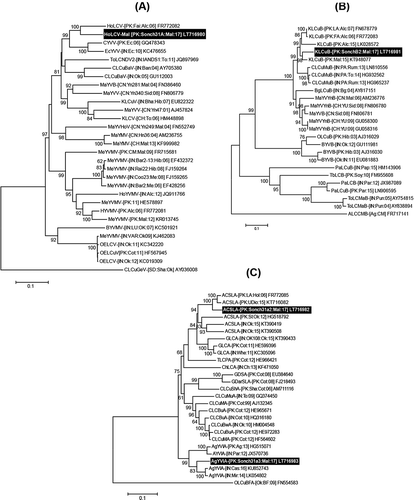
The clones Sonch31a2 and Sonch31a3 were 1365 and 1388 nt in length, respectively and were similar to the already reported alphasatellites in having an IR (TAGTATTA/C), replication-associated protein (Rep) in the virion-sense orientation and A-rich region. Both the alphasatellite clone sequences shared 86% nt sequence identity with each other. The clone Sonch31a2 showed its highest nt sequence identity (94.3%) with Ageratum conyzoides symptomless alphasatellite (ACSLA; Acc. No. KT716082) identified from nettle weed (Urtica dioica) in Pakistan (Iqbal et al. Citation2015). Whereas, the clone Sonch31a3 showed maximum nt sequence identity at 93.2% with Ageratum yellow vein India alphasatellite (AYVIA) (Acc. No. KU852743) from coffee-senna (Senna occidentalis) in India (Kumar et al. Citation2016). In the phylogenetic dendrogram, Sonch31a2 and Sonch31a3 were grouped with ACSLA and AYVIA into well-supported clades, respectively (). Thus, following the suggested species demarcation threshold >83% for alphasatellites (Mubin et al. Citation2009), Sonch31a2 and Sonch31a3 are the new isolates of ASLA (Acc. No. LT716982) and AYVIA (Acc. No. LT716983), respectively.
Recombination analysis of the begomovirus and alphasatellites
The RDP4 analysis detected two recombination events at nt coordinates 1047–1257 and 1788–2623 with HoLCV (Acc. No. FR772082) and Kenaf leaf curl virus (KLCV; Acc. No. HM448898) as major parents, whereas Hollyhock yellow vein mosaic virus (HoYVMV; Acc. No. JQ911766) and Mesta yellow vein virus (MeYVV; Acc. No. FR71568) were minor parents, respectively. The results were supported by highest P-values calculated as 1.04 × 10−12–1.08 × 10−35 and 6.80 × 10−05–7.49 × 10−13 for the two recombination events, respectively.
Both ACSLA and AYVIA isolates showed one recombination event each at nucleotide coordinates 102–509 and 138–836 with Sida yellow vein alphasatellite (SYVA; Acc. No. KM108329) and ACSLA (Acc. No. FR772085) as major parents, whereas minor parents were unknown, respectively. The recombination events were further supported by highest P-values calculated as 5.59 × 10−05–6.11 × 10−10 and 1.22 × 10−12–3.07 × 10−21, respectively.
Although M. parviflora has been reported as an alternate and/or experimental weed host for many bipartite begomoviruses, a complete begomovirus complex has never been reported from this species. To the best of our knowledge, this is the first report of a monopartite begomovirus complex identified from M. parviflora in Pakistan. The molecular characterization of diverse DNA-satellites associated with HoLCV presents evidence of the pseudo-recombination and trans-replication capabilities of HoLCV. The results further extend our knowledge about the aetiology of the begomoviruses and the interaction with their alternate weed hosts.
Acknowledgements
Fasiha Qurashi carried out this research work as part of her PhD studies. This work was supported by University of the Punjab, Lahore, Pakistan.
Additional information
Funding
References
- Akbar S, Hanif U, Ali J, Ishtiaq S. 2014. Pharmacognostic studies of stem, roots and leaves of Malva parviflora L. Asian Pac J Trop Biomed. 4:410–415.
- Barros L, Carvalho AM, Ferreira ICFR. 2010. Leaves, flowers, immature fruits and leafy flowered stems of Malva sylvestris: a comparative study of the nutraceutical potential and composition. Food Chem Toxicol. 48:1466–1472.
- Berrie LC, Rybicki EP, Rey MEC. 2001. Complete nucleotide sequence and host range of South African cassava mosaic virus: further evidence for recombination amongst begomoviruses. J Gen Virol. 82:53–58.
- Briddon RW, Brown JK, Stanley J, Zerbini M, Zhou X, Fauquet CM. 2008. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch Virol. 153:763–781.
- Briddon RW, Bull SE, Amin I, Mansoor S, Bedford ID, Rishi N, Siwatch SS, Zafar MY, Abdel-Salam AM, Markham PG. 2004. Diversity of DNA 1: a satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology. 324:462–474.
- Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. 2002. Universal primers for the PCR-mediated amplification of DNA β: a molecule associated with some monopartite begomoviruses. Mol Biotechnol. 20:315–318.
- Briddon RW, Ghabrial S, Lin N-S, Palukaitis P, Scholthof K-BG, Vetten H-J. 2012. Satellites and other virus-dependent nucleic acids. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy - Ninth Report of the International Committee on Taxonomy of Viruses. London, Waltham, San Diego: Associated Press, Elsevier Inc., p. 1209–1219.
- Briddon RW, Stanley J. 2006. Sub-viral agents associated with plant single-stranded DNA viruses. Virology. 344:198–210.
- Brown JK, Fauquet CM, Briddon RW, Zerbini M, Moriones E, Navas-Castillo J. 2012. Geminiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. London: Associated Press; 351–373.
- Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JF, Fiallo-Olivé E, Briddon RW, Hernández-Zepeda C, Idris A, et al. 2015. Revision of begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol. 160:1593–1619.
- Bull SE, Briddon RW, Markham PG. 2003. Universal primers for the PCR-mediated amplification of DNA 1: a satellite-like molecule associated with begomovirus-DNA β complexes. Mol Biotechnol. 23:83–86.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Fiallo-Olivé E, Tovar R, Navas-Castillo J. 2016. Deciphering the biology of deltasatellites from the New World: maintenance by New World begomoviruses and whitefly transmission. New Phytol. 212:680–692.
- Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 16:573–582.
- Haible D, Kober S, Jeske H. 2006. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J Virol Meth. 135:9–16.
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 11:777–788.
- Heinrich M, Barnes J, Gibbons S, Williamson E. 2012. Fundamentals of pharmacognosy and phytotherapy. Edinburgh: Elsevier; p. 326.
- Hussain K, Hussain M, Mansoor S, Briddon RW. 2011. Complete nucleotide sequence of a begomovirus and associated betasatellite infecting croton (Croton bonplandianus) in Pakistan. Arch Virol. 156:1101–1105.
- Idris AM, Hiebert E, Bird J, Brown JK. 2003. Two newly described begomoviruses of Macroptilium lathyroides and common bean. Phytopathology. 93:774–783.
- Iqbal MJ, Hussain W, Zia-Ur-Rehman M, Hameed U, Haider MS 2015. First report of Chilli leaf curl virus and associated alpha- and beta-satellite DNAs infecting nettle weed (Urtica dioica) in Pakistan. Plant Dis. 100:870–870.
- Jeske H. 2009. Geminiviruses. In: de Villiers E-M, Zur Hausen H, editors. TT viruses; The still elusive human pathogens. Vol. 331, Current topics in microbiology and immunology. Berlin: Springer Verlag; p. 185–226.
- Jeske H, Werz G. 1980. Ultrastructural and biochemical investigations on the whitefly transmitted Abutilon mosaic virus (AbMV). Phytopathol Zeitsch. 97:43–55.
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Virus taxonomy - ninth report of the International Committee on taxonomy of viruses. Associated Press; London, Waltham, San Diego. p. 1327.
- Kon T, Gilbertson RL. 2011. Two genetically related begomoviruses causing tomato leaf curl disease in Togo and Nigeria differ in virulence and host range but do not require a betasatellite for induction of disease symptoms. Arch Virol. 157:107–120.
- Kumar J, Alok A, Kumar J, Tuli R. 2016. Senna leaf curl virus: a novel begomovirus identified in Senna occidentalis. Arch Virol. 161:2609–2612.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. 21:98–102.
- Martin DP, Rybicki EP. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 16:562–563.
- Mubin M, Briddon RW, Mansoor S. 2009. Complete nucleotide sequence of chili leaf curl virus and its associated satellites naturally infecting potato in Pakistan. Arch Virol. 154:365–368.
- Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 9:e108277.
- Nawaz-ul-Rehman MS, Fauquet CM. 2009. Evolution of geminiviruses and their satellites. FEBS Lett. 583:1825–1832.
- Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology. 265:218–225.
- Paprotka T, Metzler V, Jeske H. 2010. The complete nucleotide sequence of a new bipartite begomovirus from Brazil infecting Abutilon. Arch Virol. 155:813–816.
- Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 98:13757–13762.
- Qurashi F, Sattar MN, Iqbal Z, Haider MS. 2017. First report of Cherry tomato leaf curl virus and associated DNA-satellites infesting an invasive weed Parthenium hysterophorus, from Pakistan. J Plant Pathol. 99:263–268.
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. 2005. Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Ann Rev Phytopathol. 43:361–394.
- Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol. 34:126–129.
- Wyatt SD, Brown JK. 1996. Detection of subgroup III geminivirus isolates in leaf extracts by degenerate primers and polymerase chain reaction. Phytopathology. 86:1288–1293.
- Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A. 2017. ICTV virus taxonomy profile: Geminiviridae. J Gen Virol. 98:131–133.

