Abstract
Diseased canola plants which failed to produce pods and were shorter than healthy plants in the same field were submitted to the Alberta Plant Health Laboratory in September 2016. The predominant symptom on infected plants was root rot, resembling a pink root rot. The objective of this study was to confirm the occurrence of canola pink root rot in Alberta. Two of the fungal strains isolated from symptomatic root samples were subjected to DNA barcoding that targeted three genomic regions, namely the internal transcribed spacer (ITS) region, the translation elongation factor-1α gene (EF1) and the β-tubulin gene. By similarity search (BlastN) using the resultant sequences as queries against GenBank accession sequences, both isolates were identified as Setophoma terrestris. Experiments were conducted to fulfil Koch’s postulates to demonstrate that this fungal species is a pathogen of canola. After inoculation, the two strains could cause pink root rot on canola. The fungal strains were re-isolated from the inoculated canola and their identity confirmed as S. terrestris. This is the first report of pink root rot on canola and on any Brassica host in the world.
Résumé
: En septembre 2016, des plants de canola malades qui n’avaient pas produit de cosses et qui étaient plus courts que des plants sains ont été envoyés au Alberta Plant Health Laboratory. Le symptôme prédominant était le pourridié des racines, semblable à la pourriture rose. Le but de cette étude était de confirmer l’occurrence de la pourriture rose chez le canola en Alberta. Deux des souches fongiques isolées à partir d’échantillons de racines ont été soumises au codage à barres de l’ADN qui a ciblé trois régions génomiques, à savoir la région de l’espaceur transcrit interne (ITS), le gène du facteur d’élongation 1α de la traduction (EF1) et le gène de la β-tubuline. En procédant à une recherche de similitude (Blast) avec les séquences résultantes pour les comparer aux séquences d’accessions de la GenBank, les deux isolats ont été identifiés en tant que Setophoma terrestris. Des expériences ont été menées pour valider les postulats de Koch afin de démontrer que cette espèce fongique est un agent pathogène du canola. Après inoculation, les deux souches pouvaient causer la pourriture rose des racines chez le canola. Les souches fongiques ont été isolées de nouveau à partir du canola inoculé et leur identité en tant que S. terrestris a été confirmée. Il s’agit de la première mention de pourriture rose des racines chez le canola ainsi que chez quelque Brassica que ce soit dans le monde.
Introduction
The Alberta Plant Health Lab (APHL) was established in 2016 and provides crop pest diagnostics in Alberta. During the autumn of 2016, a few canola plant samples with symptoms reminiscent of root rot were received from Castor, Alberta for pathogen identification. The pathogenic fungal species isolated from these samples were identified as Setophoma terrestris. We called the symptoms pink root rot derivatively and considered it as a new disease of canola.
Setophoma terrestris (H.N. Hansen) Gruyter, Aveskamp & Verkley is a recently named species of anamorphic fungi in the phylum Ascomycota. In 1929, Hansen identified the causal agent of onion pink root and suggested a species name Phoma terrestris (Hansen Citation1929). Subsequently, the name was changed to Pyrenochaeta terrestris by Gorenz et al. (Citation1948) since the morphology of the fruiting bodies of the pink root pathogen resembled Pyrenochaeta but differed from Phoma. Recently, de Gruyter et al. (Citation2010) reclassified this fungus to a newly developed genus Setophoma, based on the sequences of the 18S and 28S nuclear ribosomal DNA. Although S. terrestris is the current name, the two previous names are still being used in some documents and online publications.
Setophoma terrestris causes pink root rot on members in the Allium genus and other crops, such as tomato, eggplant, pepper and carrot (Sumner Citation1995). In Canada, pink root rot caused by this fungal species is commonly found on onion (McDonald Citation1994) and corn (Zhu et al. Citation2005; Pouleur et al. Citation2009). In Japan, pink root rot on squash caused by this fungal species has been reported (Ikeda et al. Citation2012). Setophoma terrestris has never been reported from canola. Therefore, this study was conducted to identify the fungal species isolated from the symptomatic canola plants and to demonstrate that the identified fungal strains could cause pink root rot on canola.
Materials and methods
Fungal isolation
Five diseased canola plants were collected from a field near Castor, Alberta, Canada in the autumn of 2016. Symptomatic root tissues were cut into 1-cm pieces and sterilized by submersion in 1% NaOCl for 1 min. The samples were rinsed three times with sterile water, and incubated on 1.5% agar supplemented with 50 µg mL−1 streptomycin at 25°C in darkness for 4 days. Hyphal tips from growing colonies were transferred onto 4% potato dextrose agar (PDA). After the colonies were established on PDA, the fungal strains were purified again by the hyphal-tip method. Two similar fungal strains were selected for further studies.
Fungal morphology
The two fungal strains were maintained on 4% PDA plates at 4°C. For sporulation and microsclerotia generation, the strains were cultured on 1.7% corn meal agar (CMA) in darkness at 25°C for 7 days before being placed under a 13-h photoperiod and 11-h darkness for another 14 days at the same temperature. Morphologies of microsclerotia, pycnidia and pycnidiospores (conidia) were investigated using a M205C dissection scope or a DM1000 microscope (Leica Microsystems, Concord, ON).
Plant inoculation
The pathogenicity of the two fungal strains was tested on seedlings of canola cultivar ‘Westar’. In an in vitro test, the root tips of 7-day-old seedlings in Petri dishes were inoculated with small pieces of PDA cut from the edge of 7-day-old fungal cultures. The dishes were incubated at room temperature for 7 days. In planta inoculation was conducted in a greenhouse. To prepare the inoculum, wheat grains (‘CDC Teal’) were soaked in water overnight and then autoclaved for 1 hour in gusseted mushroom growing bags. Small pieces of PDA of each of the two fungal strains were separately inoculated in the bags. The bags were incubated at 25°C under a 13-h photoperiod and 11-h darkness. After 21 days, the grains bearing extensive mycelia were air-dried and ground with a Micro Mill Grinder (Fisher Scientific, Ottawa, ON). The resultant powder was used as inoculum. Plastic cups (420-mL) were filled with Sunshine mix #4 soil (Sun Gro Horticulture, Vancouver, BC) and five mL of the inoculum was added to the top of the soil. Ten seeds of canola ‘Westar’ were sown in each cup and then covered with 50 mL of Sunshine mix #4 soil. The cups were kept in a greenhouse maintained at 24°C/18°C (day/night) with a 16-h photoperiod. Two experiments were conducted simultaneously, which differed in the concentration of the inoculum: the original powder or a 5× dilution (diluted with Sunshine mix #4 soil). For each experiment, the cups were arranged as randomized complete blocks with three replicates. Each block consisted of six different inocula (treatments): strain 1 inoculum (Trt1), strain 1 inoculum that had been autoclaved before air dried (CK1), strain 2 inoculum (Trt2), strain 2 inoculum that had been autoclaved before air dried (CK2), powder from autoclaved wheat grains (Grain CK) and uninoculated control (Empty CK). In addition, extra cups of Trt1 and Trt2 with both inoculum concentrations were prepared to generate materials for microscopic investigation of fungal infection.
Pathogenicity assessment
In the in vitro pathogenicity test, the inoculated plants were examined visually every day until symptoms were observed. In the in planta test, at 10 days after inoculation (dai), symptomatic roots from plants growing in the extra cups of Trt1 and Trt2 were washed with water and subjected to WGA-FITC staining following the protocol described by Manning & Ciuffetti (Citation2015). The stained samples were examined using an AxioImager M2 fluorescence microscope (Zeiss Canada, Toronto, ON) to observe the infecting mycelia. The germination rate in all cups was assessed at 10 dai. The height of each plant was measured at 14 dai. Data from each of the two inoculation rates was subjected to analysis of variance using the Microsoft Excel add-in DSAASTAT developed by Dr Onofri at the University of Perugia, Italy (http://accounts.unipg.it/~onofri/DSAASTAT). Differences between treatments were assessed using Fisher’s LSD test (P ≤ 0.05).
Fungal barcoding
Three pairs of DNA primers () were used in PCRs to amplify three fungal barcoding fragments: primer pairs ITS1/ITS4 (White et al. Citation1990) for the internal transcribed spacer (ITS) region, EF1-1018F/EF1-1620R (Stielow et al. Citation2015) for the translation elongation factor-1α gene (EF1) and T1/β-Sandy-R (Stukenbrock et al. Citation2012) for the β-tubulin gene. Primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). All PCRs were conducted in Promega PCR master mix (Promega, Madison, WI, USA) with a T100 thermal cycler (Bio-Rad Canada, Mississauga, ON). Electrophoresis was performed in 1% agarose gels stained with SYBR Green. The PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega). Other molecular techniques, if not specified, were performed according to the protocols described by Sambrook & Russell (Citation2001). Sequencing was done by the Department of Biological Sciences, University of Alberta (Edmonton, AB). Identification of fungal species was performed by similarity searches (BlastN) using the obtained sequences against the non-redundant nucleotide collection (nr/nt) hosted by the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 1. Oligonucleotide primers used in this study.
Results
Fungal isolation
The diseased canola plants collected from the field had symptoms of stunting, spindly stems and flower or pod abortion. The roots were underdeveloped and exhibited root pruning, with fewer lateral roots compared with roots of healthy plants. Black lesions similar to root rot could be observed. Fungi isolated from root samples from five plants resulted in five fungal colonies on PDA plates. One was identified as a Cladosporium sp. and a second an unidentified ascomycete-like fungus. The three remaining colonies were morphologically similar to each other and to the fungal species S. terrestris as described in the Q-bank fungi database (http://www.q-bank.eu/Fungi). Two of these colonies were purified by the hyphal-tip method. When inoculated in vitro on root tips of canola seedlings in Petri dishes, each of the two strains caused a pink discolouration of root tissues at the vicinity of the inoculation site within 3 days (data not shown), indicating infection of the root by the fungal strains. This result strengthened the assumption that the symptoms on the canola samples were caused by S. terrestris.
Fungal barcoding
The entire 544-nt ITS sequences from the two strains were identical except for a single nt at position 397 and matched the GenBank accession number KU059887 for an unidentified species of Setophoma at 99% or 100%. For both strains, the best BlastN from a known species was the ITS sequence from S. terrestris (GenBank accession number AB695295), with a 2 or 3-nt difference on the aligned 530-bp region. The 292-nt β-tubulin sequences from the two strains were different at 3 nt and BlastN matches for β-tubulin sequences from S. terrestris were 99% or 100% similar. The sequence from the first strain was identical with GenBank accession number KF252723 of S. terrestris. The 639-nt EF1 sequences from the two strains differed in a single nt. There were no EF1 sequences of S. terrestris available in the database. The BlastN for both sequences had a highest match to a sequence from Alternaria alternata, with a 96% identity based on 639-nt alignment. The sequences were submitted to GenBank with accession numbers KY561336–KY561341 for ITS, β-tubulin and EF1, respectively.
Fungal morphology
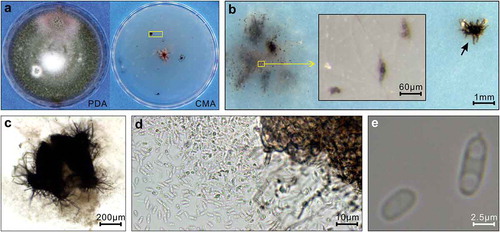
Both strains produced dark brown to pinkish colonies on PDA (). On CMA, the colonies grew as white mycelia with scattered pycnidia and microsclerotium-producing hyphal mats (). Microsclerotia were globose and brown (). The pycnidia were solitary or confluent, produced on the upper surface or submerged in agar, dark brown, globose and setose (). Conidia were released from the pycnidia on the medium and on a microscope slide (). The conidia were hyaline, aseptate, elliptical with a guttule at each end (). According to the description in Q-bank and Borema et al. (Citation2004), the observed morphological characteristics were specific to S. terrestris.
Fig. 1 (Colour online) Morphology of Setophoma terrestris isolated from canola roots. Data from strain 2 are presented. (a) Growing on 4% potato dextrose agar (PDA) for 14 days and on 1.7% corn meal agar (CMA) for 37 days. The yellow rectangle frames the area depicted in (b). (b) Microsclerotia (yellow rectangle) and a pycnidium (indicated by an arrow). The inlaid panel is the enlarged view of microsclerotia. (c) Enlarged view of the pycnidium in (b). (d) Conidia released from a pycnidium. (e) Enlarged view of conidia.
Pathogenicity assessment
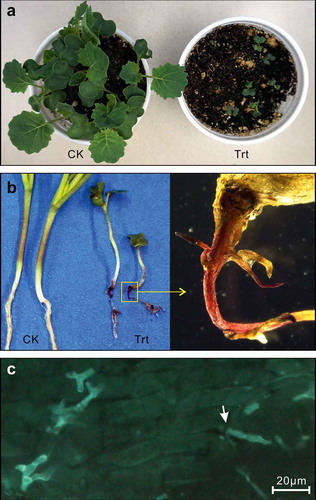
Both strains were tested for pathogenicity on canola. After germination, the inoculated plants grew slower than the uninoculated plants (). At 10 dai, infection of the entire root system could be observed on the inoculated plants (). The infected tap root became pink and shrivelled (). WGA-FITC stained samples viewed under the compound microscope showed outgrowths of hyphae from the juncture of root cells (), indicating that the fungus invaded and colonized sub-epidermal root tissues rather than simply growing across the root surface.
Fig. 2 (Colour online) Infection of canola by Setophoma terrestris isolated from canola roots. Data from strain 2 are presented. (a) Canola plants at 10 days after inoculation (dai). (b) Root symptoms at 10 dai. CK in (a) and (b), control treatment inoculated with autoclaved inoculum; Trt in (a) and (b), inoculated treatment. (c) Infected root with WGA-FITC staining. A hypha growing from the juncture of root cells is indicated by an arrow.
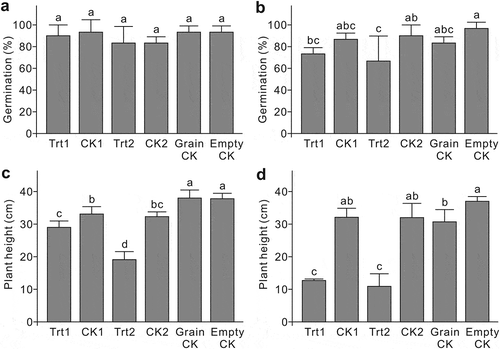
Germination and plant height were assessed at 10 and 14 dai, respectively, for both inoculum rates. In the low inoculum rate experiment, the germination ratios among treatments were similar (). At the high inoculum rate, strain 1 (Trt1) did not affect germination, compared with the CK1 (). Inoculation with strain 2 (Trt2) reduced germination, in contrast to the CK2 (). Both strains in both inoculation experiments showed a negative effect on plant height (). At the low inoculum rate (), strain 2 (Trt2) showed a more severe reduction in plant height than strain 1 (Trt1), which, along with the germination data of the high inoculation experiment (), suggests that strain 2 is more aggressive than strain 1 on the canola cultivar ‘Westar’.
Fig. 3 Pathogenicity of the two Setophoma terrestris strains isolated from canola roots, represented by germination rate and plant height after inoculation. (a) and (c) inoculated with a low-concentration inoculum. (b) and (d) inoculated with a high-concentration inoculum. Trt1 = strain 1 inoculum; CK1 = strain 1 inoculum that has been autoclaved; Trt2 = strain 2 inoculum; CK2 = strain 2 inoculum that has been autoclaved; Grain CK = powder of autoclaved wheat grains; Empty CK = uninoculated control. Means with the same letter do not differ based on Fisher’s LSD test at P ≤ 0.05 (n = 3).
Root samples were collected from the inoculated canola seedlings at 14 dai and subjected to fungal isolation. Four fungal strains were obtained, with two from Trt1 and two from Trt2-infected roots. The morphologies of these four strains were identical to each other and to strain 1 and strain 2, confirming that the same fungus was re-isolated from the inoculated, symptomatic plant and thus fulfilling Koch’s postulates (Agrios Citation2005).
Discussion
The two fungal strains causing pink root rot on canola were identified as S. terrestris using the ITS and the β-tubulin gene sequences following queries in BlastN, with similarities of 99–100%. However, BlastN using the EF1 sequences did not generate matches with S. terrestris. Since the EF1 region investigated in the current study is recently reported (Stielow et al. Citation2015), there are no S. terrestris sequences of the corresponding region available in the database. This EF1 region is located downstream of the widely used EF1 region (O’Donnell et al. Citation1998) and has been demonstrated to be a promising region for fungal DNA barcoding (Stielow et al. Citation2015).
ITS and β-tubulin sequences showed small polymorphisms (1–3 nt) between the two strains. This indicates that there is some genetic variation among the S. terrestris field isolates. The two strains also differed in their pathogenicity, with strain 2 being more aggressive than strain 1, as indicated by differences in germination and plant height of canola plants inoculated separately with one of the two strains. Whether the differences in pathogenicity are related to the genetic variation is unknown and requires further studies on a population or phylogenetic level.
Setophoma terrestris is not always pathogenic even if originating from plant samples because it is a common soil inhabitant (Gorenz et al. Citation1949). Thus, in this study, we tested pathogenicity of the isolated strains on canola by fulfilling Koch’s postulates. The data demonstrated the pathogenic nature of S. terrestris on canola. The extent to which the disease may become prevalent if field and weather conditions are appropriate is unknown.
It is known that pink root rot is worse when onions are planted after cereals (Sumner Citation1995; Nischwitz & Dhiman Citation2012). In Alberta, crop rotations heavily favour cereals and canola. According to the sample submitted, the infected field was on a 3-year rotation: barley followed by triticale and then canola. This rotation (two cereal crops preceding canola) may have increased the opportunity for populations of the pink root rot pathogen to build and reach the threshold necessary to appear as a root rot pathogen on canola. The distribution of the pathogen in Alberta and the extent of the disease in other regions are unknown.
Additional information
Funding
References
- Agrios GN. 2005. Plant pathology. 5th ed. San Diego (CA): Elsevier Academic Press; p. 26–27.
- Borema GH, de Gruyer J, Noodeloose ME, Hamers MEC. 2004. The methods used for differentiation and identification. In: Borema GH, de Gruyer J, Noordeloos ME, Hamers MEC, editors. Phoma identification manual: differentiation of specific and intra-specific taxa in cultures. Cambridge (MA): CABI Publishing; p. 14–18.
- de Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW. 2010. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 102:1066–1081.
- Gorenz AM, Larson RH, Walker JC. 1949. Factors affecting pathogenicity of pink root fungus of onions. J Agric Res. 78:1–18.
- Gorenz AM, Walker JC, Larson RH. 1948. Morphology and taxonomy of the onion pink-root fungus. Phytopathology. 38:831–840.
- Hansen HN. 1929. Etiology of the pink-root disease of onions. Phytopathology. 19:691–704.
- Ikeda K, Kuwabara K, Urushibara T, Soyai P, Miki S, Shibata S. 2012. Pink root rot of squash caused by Setophoma terrestris in Japan. J Gen Plant Pathol. 78:372–375.
- Manning VA, Ciuffetti LM. 2015. Necrotrophic effector epistasis in the Pyrenophora tritici-repentis-wheat interaction. PLoS One. 10:e0123548.
- McDonald MR. 1994. Pink root. In: Howard RJ, Garland JA, Seaman WL, editors. Diseases and pests of vegetable crops in Canada. Ottawa (ON): The Canadian Phytopathological Society and the Entomological Society of Canada; p. 281–282.
- Nischwitz C, Dhiman C. 2012. Pink root of onion. Utah Pests Fact Sheet, PLP-017. Utah State University Extension and Utah Plant Pest Diagnostic Laboratory. http://extension.usu.edu/files/publications/factsheet/PLP-017-July2012.pdf.
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA. 95:2044–2049.
- Pouleur S, van Herk C, Tenuta A. 2009. Survey of red root rot of corn in Ontario in 2008. Can Plant Dis Surv. 89:70–73.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Stielow JB, Lévesque CA, Seifert KA, Meyer W, Iriny L, Smits D, Renfurm, R, Verkley, GJM, Groenewald, M, Chaduli, D, et al. 2015. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 35:242–263.
- Stukenbrock EH, Quaedvlieg W, Javan-Nikhah M, Zala M, Crous PW, McDonald BA. 2012. Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola). Mycologia. 104:1397–1407.
- Sumner DR. 1995. Pink root. In: Schwartz HF, Mohan SK, editors. Compendium of onion and garlic diseases. St. Paul (MN): American Phytopathological Society Press; p. 12–13.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. London (UK): Academic Press; p. 315–322.
- Zhu X, Reid LM, Woldemariam T, Tenuta A, Lachance P, Pouleur S. 2005. Survey of corn diseases and pests in Ontario and Québec in 2004. Can Plant Dis Surv. 85:31–34.
