Abstract
Villosiclava virens is an ascomycete fungus (anamorph: Ustilaginoidea virens) recognized as one of the most economically important pathogens on rice in China that causes false smut disease. In our study, the V. virens cyp51 gene encoding the target protein for fungicide resistant to tebuconazole was cloned. Sequencing showed that fungal mutant strains resistant to tebuconazole harboured the T409C mutation in cyp51. A simple and rapid allele-specific PCR assay was developed to detect the T409C mutation. Among the isolates tested, a unique 391-bp fragment was amplified from resistant mutant strains with primer pair Uv137F1/Uv137DR, whereas no product was amplified from all sensitive isolates. Our study demonstrated that this allele-specific PCR assay is a rapid, simple and reliable method for identification of V. virens mutants resistant to tebuconazole.
Résumé
Villosiclava virens est un ascomycète (forme imparfaite: Ustilaginoidea virens) qui cause le faux charbon et qui est reconnu comme un des agents pathogènes qui a la plus grande importance économique en ce qui a trait au riz en Chine. Dans notre étude, le gène cyp51 de V. virens, qui code la protéine cible pour le fongicide tébuconazole, a été cloné. Le séquençage a montré que les mutants fongiques résistants au tébuconazole hébergeaient la mutation T409C dans le gène cyp51. Une analyse simple et rapide par PCR spécifique de l’allèle a été conçue pour détecter la mutation T409C. Parmi les isolats testés, un fragment unique de 391 pb a été amplifié à partir de mutants résistants avec une paire d’amorces Uv137F1/Uv137DR, alors qu’aucun produit issu de tous les isolats sensibles n’a été amplifié. Notre étude a démontré que cette PCR spécifique de l’allèle est une méthode rapide, simple et fiable qui permet d’identifier les mutants de V. virens résistants au tébuconazole.
Introduction
Rice false smut, caused by Villosiclava virens (Nakata) E. Tanaka & C. Tanaka (anamorph: Ustilaginoidea virens), is a fungal disease infecting rice spikelets (Tanaka et al. Citation2008), specifically rice stamen filaments (Tang et al. Citation2013; Hu et al. Citation2014). Cooke (Citation1878) first isolated and named the pathogen based on ‘a rice smut’ collected in India. Historically, rice false smut was considered to be a minor disease. Since the 1980s, with expanded planting of hybrid rice cultivars with large and compact panicles, increased use of fertilizers, and high levels of irrigation, it has become one of the most important diseases in China (Zhu, Xu et al. Citation2007). In Fujian Province of China, the number of false smut balls per panicle is usually 1–10, and sometimes up to 30–50 in the field (Yang et al. Citation2013). In addition to reducing rice yield and grain quality (Jiang et al. Citation2009; Yang et al. Citation2012), the disease is poisonous to both humans and animals owing to the production of a large quantity of mycotoxins (Koiso et al. Citation1998; Joullie et al. Citation2011).
Sterol demethylation inhibitors (DMIs), including triazoles, imidazoles, piperazines, pyrimidines and pyridines (Hamamoto et al. Citation2000), are a major class of fungicides used in agriculture, targeting the haeme iron of cytochrome P450 sterol 14α-demethylase (CYP51) to interfere with the biosynthesis of ergosterol (Yoshida & Aoyama Citation1987). However, because of their site-specific mode of action, consecutive applications of DMIs have led to the emergence and development of resistance in a number of plant pathogenic fungi (Schnabel et al. Citation2004; Ulrich Citation2014; Rallos & Baudoin Citation2016).
Tebuconazole, a DMI fungicide developed in the late 1990s by Bayer, has been registered and widely applied in more than 50 countries to more than 60 crops as a broad-spectrum antifungal compound (Gu Citation2010). However, the excessive use of tebuconazole had provoked reduced sensitivity in several important plant pathogens, such as Botryosphaeria dothidea (Fan et al. Citation2013), Lasiodiplodia theobromae (Pereira et al. Citation2012), and Colletotrichum gloeosporioides (Chen et al. Citation2013). At present, tebuconazole is used to control many rice diseases in China, including false smut (Dou et al. Citation2015), sheath blight (Wang et al. Citation2007) and blast disease (Liu et al. Citation2013). Although the resistance of V. virens to tebuconazole has not been reported in the field, with the extensive use of the fungicide, the risk of resistance of V. virens to tebuconazole is increasing. Five laboratory tebuconazole-resistant mutants were obtained by the fungicide-taming method in our previous study. In the present study, we cloned and sequenced the cyp51 gene of sensitive isolates and resistant mutants. Based on the sequences of the cyp51, we developed an allele-specific PCR assay to detect the T409C mutation involved in tebuconazole resistance. This assay represents a useful tool for resistant surveillance of V. virens to tebuconazole.
Materials and methods
Villosiclava virens isolates and false smut balls used in this study
Fifteen wild-type isolates, isolated from Fujian Province of China, were sensitive to tebuconazole (). Tebuconazole-resistant mutants F10-338B, F10-338C and F10-338D () were derived from the sensitive isolate F10-338, and tebuconazole-resistant mutants JY13107C and JY13107D () were derived from the sensitive isolate JY13107, through the fungicide-taming method (Gu et al. Citation2010).
Table 1. Fifteen wild-type isolates and five tebuconazole-resistant mutants of Villosiclava virens used in this study.
False smut balls (1–20) were obtained by artificial inoculation of each of the 20 isolates () onto rice cultivar ‘LYP9ʹ, which was provided by the Rice Research Institute of Fujian Academy of Agriculture Science. The conditions for inoculation were according to those described by Yang et al. (Citation2011). The symptoms of rice false smut are shown in .
PCR amplification and sequencing
PCR amplification reactions were performed in a 25 µL reaction volume containing 0.5 units of KOD-Plus-Neo (Toyobo, Shanghai, China), 2.5 µL 10×PCR buffer, 0.1 mM of each dNTP, 0.75 mM MgSO4, 0.15 µM of each primer, and 50 ng of template DNA. Amplification was carried out in a C1000TM thermal cycler (Bio-Rad, CA) with the following profile: 94°C for 3 min, 36 cycles at 98°C for 10 s, 59.6°C (S-4 and A-3) or 57.2°C (S-2 and A-2) for 30 s, and 68°C for 80 s (S-4 and A-3) or 50 s (S-2 and A-2), and a final extension at 68°C for 10 min. All amplified PCR products were purified with a gel extraction kit (Aidlab Biotech, Beijing, China), then ligated into a vector using the zero background pTOPO-Blunt simple cloning kit (Omega Biotech, Guangzhou, China), and sequenced with vector primers at Sangon Biotech (Shanghai, China).
Cloning the cyp51 gene from V. virens
Genomic DNA of seven isolates (sensitive isolates F10-338 and JY13107, and resistant mutants F10-338B, F10-338C, F10-338D, JY13107C and JY13107D), was extracted from mycelia using the HP Plant DNA Kit (Omega Biotech) according to the manufacturer’s instructions. The complete cyp51 sequences were amplified with a pair of PCR primers S-4 and A-3 (). The conditions and systems for PCR amplification were described above. The resulting DNA sequences were analysed using the DNAMAN5.2.2.0 software package (Lynnon Biosoft, Quebec, Canada).
Table 2. Primers used in this study.
RNA extraction and cDNA synthesis of the cyp51 gene
The seven isolates were grown in 250 mL potato sucrose broth (PSB) at 28°C for 5 days on a 200 rpm orbital shaker. Total RNA was extracted from mycelia using a fungal RNA kit (Omega Biotech). The cDNA was synthesized using the ReverTra Ace -α- RT kit employing the oligo(dT)18 primer (Toyobo). The cDNA was amplified with a pair of PCR primers S-2 and A-2 (). The conditions and systems for PCR amplification were described above. The resulting RNA sequences were analysed using the DNAMAN5.2.2.0 software package (Lynnon Biosoft, Quebec, Canada). The entire experiment, starting with cloning the cyp51 gene, was repeated twice.
AS-PCR marker: primer design
DNAMAN software was used to identify the mutation site between sensitive isolates and resistant mutants in the coding region of cyp51. Based on the single-point mutation at the codon position 409 (T409C) in the cyp51 gene of resistant mutants, we designed four pairs of allele-specific primers for detection of the point mutation. A nucleotide G at 3ʹ-end of the reverse primer Uv137AR was designed to match the C at codon 409 in resistant mutants. To improve the specificity, the second nucleotide T at 3ʹ-end of the reverse primer Uv137AR was changed to A, G and C in primers Uv137BR, Uv137CR and Uv137DR, respectively (). All allele-specific PCR primers employed the same forward primer Uv137F1 ().
Allele-specific PCR optimization
PCR amplifications were carried out using four primer pairs. Reactions were performed in a final volume of 25 µL containing 1.25 units of Taq DNA polymerase (Tiangen Biotech, Beijing, China), 2.5 µL 10×Taq buffer, 0.1 mM of each dNTP, 0.15 µM of each primer, and 50 ng of template DNA. PCR was optimized by varying the annealing temperature (Ta) (44°C–56°C). The PCR profile was 94°C for 3 min, followed by 36 cycles at 98°C for 10 s, gradient annealing temperatures from 56.0°C, 55.3°C, 54.0°C, 51.6°C, 48.7°C, 46.4°C, 44.8°C to 44.0°C for 30 s, 68°C for 45 s, and a final extension at 68°C for 10 min.
Sensitivity of allele-specific PCR
The concentration of genomic DNA of the resistant mutant F10-338D was measured using a Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA) at 260 nm, adjusted to 600 ng µL−1, and serially diluted 10 times with high purity distilled water to concentrations of 60, 6, 0.6, 0.06, 0.006 and 0.0006 ng µL−1. The precision was determined by analysing the lowest detection limit demonstrated by allele-specific PCR.
Validating the T409C mutation using allele-specific PCR
To determine the specificity of the optimized allele-specific PCR, 20 isolates, including 15 sensitive isolates and five resistant mutants, were tested as described above. The PCR conditions and profile were the same as above except that the annealing temperature was 51.6°C. Direct PCR sequencing of positive bands was performed by Sangon Biotech.
Application of allele-specific PCR
The usefulness of allele-specific PCR was further evaluated using 20 false smut balls. These false smut balls were ground rapidly in liquid nitrogen and genomic DNA was extracted using the HP Plant DNA Kit (Omega Biotech) and tested using the optimized allele-specific PCR described above.
Results
Cloning and sequence analysis of the cyp51 gene
The entire cyp51 gene from seven isolates was amplified. The gene was 1827 bp in length and contained two introns. The full-length cDNA was 1587 bp and encoded a putative polypeptide of 528 amino acids. The gene sequences of cyp51 from isolate F10-338 and F10-338D were submitted to the GenBank database (accession no. KX609627 and MF196892). Alignment of the cyp51 gene cDNA sequences from sensitive isolates and resistant mutants showed a thymine (T) – cytosine (C) mutation at position 409 () that was correlated with tebuconazole resistance. This mutation led to a replacement of tyrosine 137 by histidine.
Optimum allele-specific PCR conditions
An allele-specific PCR assay was designed to specifically detect the T409C mutation using the genomic DNA templates. We tested the specificity of four pairs of primers by adjusting the annealing temperature with sensitive isolate F10-338 and resistant mutant F10-338D. Two independent experiments showed that the optimal annealing temperature was 51.6°C and the optimal primer pair was Uv137F1/Uv137DR. A 391-bp fragment was amplified from resistant mutant F10-338D whereas no amplification was obtained from sensitive isolate F10-338. Thus, in our study, PCR with the Uv137F1/Uv137DR primer for the T409C mutation of the cyp51 gene specifically amplified a 391-bp fragment from resistant mutants at an annealing temperature of 51.6°C.
Sensitivity of allele-specific PCR
To evaluate the sensitivity of the allele-specific PCR assay, genomic DNA of resistant mutant F10-338D was diluted 10 times. The allele-specific PCR assay detected the T409C mutation down to 0.006 ng µL−1 genomic DNA. Thus, a concentration of 0.006 ng µL−1 represented the limit of detection for this assay. The allele-specific PCR assay for the lowest limit of detection is shown in .
Validating the T409C mutation using allele-specific PCR
In order to validate the specificity and accuracy of the allele-specific PCR, 20 isolates, including 15 sensitive isolates and five resistant mutants (), were evaluated using optimized allele-specific PCR. The results revealed that a 391-bp fragment was amplified from resistant mutants, whereas no product was amplified from sensitive isolates (). Sequencing results were 100% consistent with the allele-specific PCR detecting the T490C mutation in V. virens. This demonstrated that the allele-specific PCR developed in this study was a reliable and specific method to distinguish resistant mutants from sensitive isolates.
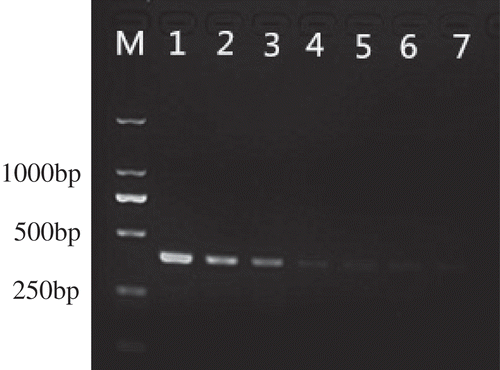
Fig. 4 Allele-specific PCR amplification products from genomic DNA extracted from eight tebuconazole-sensitive isolates and five tebuconazole-resistant mutants of V. virens. M, 2-kb DNA ladder (Takara Bio); lane 1, negative control; lanes 2–9, isolates F10-338, JY13107, JY1601, NH13018, NH13047, SX13023, SH13002 and SH13003; Lanes 10–14, isolates F10-338B, F10-338C, F10-338D, JY13107C and JY13107D.

Application of allele-specific PCR
Twenty false smut balls were tested for the T409C mutation using optimized allele-specific PCR. Of the 20 false smut balls tested, a 391-bp band was obtained only from five false smut balls derived from resistant mutants but not from any false smut balls derived from sensitive isolates (). The assay could be used to detect resistance of V. virens to tebuconazole in planta.
Fig. 5 Allele-specific PCR amplification products from genomic DNA extracted from false smut balls. M, 2-kb DNA ladder (Takara Bio); lane 1, negative control; lanes 2–6, false smut balls derived from tebuconazole-resistant mutants; lanes 7–18, false smut balls derived from tebuconazole-sensitive isolates.

Discussion
To date, the allele-specific PCR method has been successfully used for the detection of fungicide resistance in numerous plant fungal pathogens (Yin & Xiao Citation2013). In this study, we successfully established an allele-specific PCR assay to detect the T409C mutation correlated with tebuconazole resistance in V. virens. Successful allele-specific PCR assays require extensive optimization of primers and amplification conditions. Artificially introducing a mismatched base, together with the mutant base at the 3ʹ end of the primer, has been shown to produce a significant increase in specificity for allele-specific PCR at the optimum annealing temperature (Hayashi et al. Citation2004; Zhu, Zhang et al. Citation2007; Yin & Xiao Citation2013). In our study, when the T in the second nucleotide position of the 3ʹ end of the primer Uv137AR was changed to C in the primer Uv137DR, the specificity of the primer Uv137DR was improved remarkably. Our study demonstrated that the optimal base change at the second nucleotide of the 3ʹ end of a primer could only be determined after each possible base change has been tested. Further, false smut balls obtained from artificial inoculation were tested using the allele-specific PCR assay. The results showed that a unique and bright 391-bp band was only amplified from five false smut balls derived from resistant mutants, as expected. Therefore, the allele-specific PCR assay may be used to directly detect the resistance of false smut balls to tebuconazole in the field in the future. This assay may contribute to monitoring the resistance of V. virens to tebuconazole in the field.
The allele-specific PCR assay that was developed here has more advantages than currently available methods. First, the allele-specific PCR method is a simple, rapid, and easy to perform test compared with traditional biological methods which are labour intensive and time-consuming. Second, the allele-specific PCR method has high accuracy that can effectively discriminate between sensitive isolates and resistant isolates. In addition, this assay does not require additional post-PCR product preparations, such as the DNA purification step in the DNA sequencing method. Further, allele-specific PCR shows high sensitivity for detection of the resistant mutant with approximately 0.006 ng µL−1 genomic DNA. Finally, this allele-specific PCR method has wide applicability, suitable for detecting V. virens mutants from not only fungal cultures but also rice false smut balls.
Tebuconazole is now considered the primary fungicide to control rice false smut (Wang Citation2015). Although resistant isolates have not been reported in the field, the intensive use of tebuconazole to control rice false smut will likely lead to tebuconazole resistance in V. virens populations in the future. Allele-specific PCR provides rapid, simple, and inexpensive detection of the T409C mutation linked with tebuconazole resistance. This assay can monitor the occurrence and development of resistance. Therefore, this method has great significance by providing an early warning of resistance in V. virens populations and establishing a disease management programme to prolong fungicide effectiveness and delay resistance development.
Additional information
Funding
References
- Chen R, Shi HJ, Wu HM, Xu ZH, Zhang CQ. 2013. Resistance of Colletotrichum gloeosporioides causing grape ripe rot to thiophanate-methyl and tebuconazole in Zhejiang. J Fruit Sci. 30:665–668.
- Cooke MC. 1878. Some extra-European fungi. Grevillea. 7:13–15.
- Dou JX, Geng ZY, Liu CH, Li YT. 2015. Determination of combined effect of difenoconazole and tebuconazole and mixed rice false smut. Chin Agric Sci Bull. 31:185–189.
- Fan K, Qu JL, Li LG, Wang T, Qin X, Li XJ. 2013. Study on baseline sensitivity of Botryosphaeria dothidea to tebuconazole and the biological characteristics of tebuconazole-resistant mutants. J Fruit Sci. 30:650–656.
- Gu CB, 2010. Study on the resistance of Fusarium oxysporum f. sp. fragariae to carbendazim and tebuconazole [dissertation]. Taian (China): Shandong Agricultural University.
- Gu CB, Jiang LL, Wang KY, Shi XB, Duan HM, Lin CH. 2010. Induction and characteristics of Fusarium oxysporum f. sp. fragariae ZY-W resistant to tebuconazole. Sci Agric Sin. 43:2897–2904.
- Hamamoto H, Hasegawa K, Lee YJ, Makizumi Y, Akutsu K, Hibi T. 2000. Tandem repeat of a transcriptional enhancer upstream of the sterol 14α-demethylase gene (CYP51) in Penicillium digitatum. Appl Environ Microbiol. 66:3421–3426.
- Hayashi K, Hashimoto N, Daigen M, Ashikawa I. 2004. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor Appl Genet. 108:1212–1220.
- Hu ML, Luo LX, Wang S, Liu YF, Li JQ. 2014. Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles. Eur J Plant Pathol. 139:67–77.
- Jiang ZQ, Jiang ZC, Jiang ZH, Zhang GJ. 2009. Effect of false smut ball number on yield of rice. North Rice. 39:53–54.
- Joullie MM, Berritt S, Hamel E. 2011. Structure-activity relationships of ustiloxin analogues. Tetrahedron Lett. 52:2136–2139.
- Koiso Y, Morisaki N, Yamashita Y, Mitsui Y, Shirai R, Hashimoto Y, Iwasaki S. 1998. Isolation and structure of an antimitotic cyclic peptide, ustiloxin F: chemical interrelation with a homologous peptide, ustiloxin B. J Antibiot (Tokyo). 51:418–422.
- Liu Y, Sang HX, Wang JS, Yu YH, Song CY, Ma XH, Yu SZ. 2013. Study on the optimum application period of trifloxystrobin·tebuconazole 75% WG against main rice diseases in coastal region. World Pestic. 35:59–61.
- Pereira AVDS, Martins RB, Michereff SJ, Silva MBD, Câmara MPS. 2012. Sensitivity of Lasiodiplodia theobromae from Brazilian papaya orchards to MBC and DMI fungicides. Eur J Plant Pathol. 134:489–498.
- Rallos LE, Baudoin AB. 2016. Co-occurrence of two allelic variants of CYP51 in Erysiphe necator and their correlation with over-expression for DMI resistance. PLoS One. 11:e0148025.
- Schnabel G, Bryson PK, Bridges WC, Brannen PM. 2004. Reduced sensitivity in Monilinia fructicola to propiconazole in Georgia and implications for disease management. Plant Dis. 88:1000–1004.
- Tanaka E, Ashizawa T, Sonoda R, Tanaka C. 2008. Villosiclava virens gen. nov., comb. nov., teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon. 106:491–501.
- Tang YX, Jin J, Hu DW, Yong ML, Xu Y, He LP. 2013. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 62:1–8.
- Ulrich G. 2014. Assessment of selection and resistance risk for demethylation inhibitor fungicides in Aspergillus fumigatus in agriculture and medicine: a critical review. Pest Manag Sci. 70:352–364.
- Wang F 2015. Study on genetic diversity and DMI fungicide resistance molecular mechanism of Villosiclava virens [dissertation]. Wuhan (China): Huazhong Agricultural University.
- Wang HC, Zhou MG, Zhang YJ, Cheng CJ, Wang JX. 2007. Fungicidal activity of tebuconazole against Rhizoctonia solani and its application to rice. Chin J Pestic Sci. 9:357–362.
- Yang LM, Chen L, Juan XU, Liu JC, Ding KJ. 2012. Estimation of yield loss caused by rice false smut. J Anhui Agric Univ. 39:474–477.
- Yang XJ, Lin TB, Ruan HC, Shi NN, Du YX, Gan L, Chen FR. 2013. Effect of false smut on yield of rice and disease resistance detection of new rice varieties. Chin J Topical Crops. 34:1309–1313.
- Yang XJ, Wang ST, Ruan HC, Shi NN, Gan L, Chen FR. 2011. Artificial inoculation techniques of rice false smut in greenhouse. J Plant Protec. 38:395–400.
- Yin YN, Xiao CL. 2013. Molecular characterization and a multiplex allele-specific PCR method for detection of thiabendazole resistance in Penicillium expansum from apple. Eur J Plant Pathol. 136:703–713.
- Yoshida Y, Aoyama Y. 1987. Interaction of azole antifungal agents with cytochrome P-450 14DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol. 36:229–235.
- Zhu HC, Xu XP, Xiao GY, Yuan LP, Li BJ. 2007. Enhancing disease resistances of Super Hybrid Rice with four antifungal genes. Sci China C Life Sci. 50:31–39.
- Zhu LX, Zhang ZW, Liang D, Jiang D, Wang C, Du N, Zhang Q, Mitchelson K, Cheng J. 2007. Multiplex asymmetric PCR-based oligonucleotide microarray for detection of drug resistance genes containing single mutations in Enterobacteriaceae. Antimicrob Agents Chemother. 51:3707–3713.