Abstract
Field isolates of Stemphylium spp. collected from lentil and nine other western Canadian crops were compared to the ex-type isolate of Stemphylium botryosum by morphology and by molecular phylogenetic analysis based on the internal transcribed spacer (ITS) and the glyceraldehyde-3-phosphate dehydrogenase gene regions. Sequence data of these genes for six other Stemphylium spp. obtained from GenBank were also included in the phylogenetic analysis. Morphology of colonies and morphometry of conidia agreed with previous descriptions of S. botryosum, but did not correlate with the clustering observed in the phylogenetic analysis. Sequence data revealed that S. botryosum is one of probably two Stemphylium spp. involved in the development of stemphylium blight on Canadian lentil, and is associated with other hosts such as faba bean, soybean, bean, pea, alfalfa, coriander, caraway and fenugreek. More than one validly described Stemphylium sp. was associated with the second cluster of isolates causing stemphylium blight, indicating that an extensive taxonomic review of the genus Stemphylium is required before this second stemphylium blight pathogen can be identified by name.
Résumé
Des isolats de souches naturelles de Stemphylium spp., collectés dans l’Ouest canadien sur des lentilles et neuf autres cultures, ont été comparés morphologiquement à l’ancien type d’isolat de Stemphylium botryosum ainsi que par analyse phylogénétique en se basant sur l’espaceur transcrit interne (ITS) et les régions du gène de la glycéraldéhyde–3-phosphate déshydrogénase. Les données relatives aux séquences de ces gènes pour six autres Stemphylium spp. obtenus de la GenBank ont également été incluses dans l’analyse phylogénétique. La morphologie des colonies et la morphométrie des conidies concordaient avec les descriptions précédentes de S. botryosum, mais ne correspondaient pas au regroupement observé lors de l’analyse phylogénétique. Les données relatives aux séquences ont révélé que S. botryosum est probablement un de deux Stemphylium spp. impliqués dans le développement de la brûlure stemphyllienne chez la lentille au Canada et qu’il est associé à d’autres hôtes tels que la féverole, la fève de soja, la fève, le pois, la luzerne, la coriandre, le carvi et le fenugrec. Plus d’un Stemphylium spp. correctement décrit a été associé au deuxième regroupement d’isolats causant la brûlure stemphyllienne, ce qui indique qu’une révision taxinomique extensive du genre Stemphylium est requise avant que nous puissions nommer adéquatement ce deuxième agent pathogène causant la brûlure stemphyllienne.
Introduction
Stemphylium botryosum Wallroth is the species commonly associated with stemphylium blight on lentil (Rahman et al. Citation2010). Many species have been designated in Stemphylium since Wallroth (Citation1833) originally described this genus, naming the type species of the genus S. botryosum. Unfortunately, the description of S. botryosum by Wallroth relied on a limited number of traits restricted to morphological features of the mycelium and the shape and colour of spores (Wallroth Citation1833). Wiltshire (Citation1938) published detailed descriptions of Stemphylium spp. based on conidium and conidiophore morphology including measurements of conidia. He also associated S. botryosum with Pleospora herbarum Rabenhorst as the teleomorph. Simmons (Citation1967) addressed the problem of misclassification and misidentification among pathogenic species in the genera Stemphylium, Alternaria and Ulocladium and included descriptions and discussion of similarities among them. He emphasized that only for S. botryosum was there solid evidence for its sexual morph in P. herbarum. The association of Pleospora spp. with at least five other Stemphylium species – S. vesicarium, S. majusculum (Simons), S. triglochinicola (Sutton & Pirozynski), S. lancipes (Ellis & Everhart) and S. globuliferum (Vestergren) – was subsequently made, and it was shown that S. globuliferum had been commonly misidentified as S. sarciniforme or S. botryosum (Simmons Citation1969). Finally in 1985, the anamorph–teleomorph associations were corrected to S. herbarum–P. herbarum, S. botryosum–P. tarda, and S. alfalfae–P. alfalfa (Simmons) (Simmons Citation1985).
More recently, studies to differentiate species in the genus Stemphylium have repeatedly demonstrated overlap in morphological characters, making the identification and description of species difficult, as a result of which molecular phylogenetic analysis has been increasingly used as an additional tool to classify species (Câmara et al. Citation2002; Wang et al. Citation2010). While the nuclear rDNA internal transcribed spacer (ITS) is considered the barcoding region for fungi (Crous et al. Citation2014), glyceraldehyde-3-phosphate dehydrogenase (gpd), elongation factor 1-alpha (EF-1 alpha) and the intergenic spacer between vacuolar membrane ATPase catalytic subunit A (vmaA) and vacuole protein sorting A (vpsA) (vmaA-vpsA) have also been used for evolutionary reconstruction of Stemphylium species (Inderbitzin et al. Citation2009; Pei et al. Citation2011; Deng et al. Citation2014; Kurose et al. Citation2015; Graf et al. Citation2016). Although the ITS sequence is the most frequently used locus in phylogenetic studies of fungi and is estimated to correctly identify a species with a probability of 73% (Crous et al. Citation2014), it is often complemented with other gene sequences, such as the gpd region, for the exploration of phylogenetic relationships among Stemphylium spp. (Wang et al. Citation2010).
In Saskatchewan, stemphylium blight has developed into a common disease of widely grown lentil, and as a consequence, resistance breeding to this disease has become one of the main breeding objectives. This study was initiated to identify the causal agent(s) of stemphylium blight on lentil and to compare those to isolates from other crops and the ex-type of S. botryosum. It was hypothesized that S. botryosum was causing stemphylium blight of lentil.
Materials and methods
Isolates
Fourteen Canadian field isolates originating from stemphylium blight lesions on lentil, two from lentil in Bangladesh, one from lentil in Pakistan and 14 isolates from eight other host plants were selected from the culture collection of the Pulse Crop Pathology Research Group of the Crop Development Centre, University of Saskatchewan, based on different geographic origin and different soil and climatic zones (in case of more than one isolate per plant species) (). Additionally, the ex-type specimen of S. botryosum DAOM 195299 obtained from the Canadian Collection of Fungal Cultures in Ontario, Canada, and a Canadian field isolate of Alternaria sp. were included.
Table 1. Field isolates of Stemphylium spp., ex-type specimen of S. botryosum DAOM 195299 and Alternaria sp. selected for morphological characterization and phylogenetic analysis based on rDNA internal transcribed spacer (ITS) and glyceraldehyde–3 phosphate dehydrogenase (gpd) region sequences.
Morphological and morphometric isolate description
Morphological descriptions of isolates was restricted to culture morphology and spore size because these characters were deemed to have the highest potential to differentiate among groups based on previously published descriptions (e.g. Wallroth Citation1833; Wiltshire Citation1938; Simmons Citation1969).
For descriptions of colony colour, shape and texture, the 31 field isolates and the ex-type of S. botryosum were cultured on potato dextrose agar plus V8® vegetable juice (V8-PDA) medium (10 g potato dextrose agar [BD DifcoTM, Thermo Fisher Scientific], 10 g granulated agar [Thermo Fisher Scientific], 3 g CaCO3 [Thermo Fisher Scientific], 150 mL V8® vegetable juice [Campbell Soup Company], 850 mL distilled water) and incubated for 7 days at 25°C under continuous light (Câmara et al. Citation2002).
Conidia were arbitrarily picked using a dissecting needle from 7-day old cultures, were placed in distilled water on glass slides and 50 conidia were measured in a Carl Zeiss compound microscope at 20× magnification. Representative pictures of conidia were taken with the ZEN Carl Zeiss camera. Conidial length and width were measured using the ZEISS ZEN Digital Imaging for light microscopy micrometer calibrated at 20×. Data were analysed with the mixed model procedure in SAS 9.4 software (SAS Institute Inc., Cary, NC) specifying isolates as fixed and replicate conidia as random factors. The length and width of conidia of field isolates were compared with those of the ex-type of S. botryosum by single-degree-of-freedom contrasts.
Molecular phylogenetic analysis
All field isolates, the ex-type DAOM 195299 and the isolate of Alternaria sp. were grown on 50% potato dextrose agar (PDA) (14.6 g of potato dextrose agar [BD DifcoTM, Thermo Fisher Scientific], 750 mL distilled water) as monoconidial cultures. For each isolate, a mycelial plug was transferred into liquid potato dextrose medium (4 g potato extract, 20 g dextrose [both Thermo Fisher Scientific], 876 mL distilled water) which was incubated at 23°C under continuous light and continuous shaking at 113 rpm. Mycelia were filtered through sterile miracloth after 3 d of incubation, transferred into 2 mL vials and frozen at −80°C for 24 h, before being lyophilized for another 24 h. Lyophilized mycelia were pulverized by introducing two to three glass pearls into each tube and shaking them for 5 min in a mechanical shaker at approximately 1000 rpm. Genomic DNA was extracted with the DNeasyTM Plant Mini Kit-QIAGEN following the manufacturer’s instructions (QIAGEN Citation2012), stored in deionized RNA-free water at −20°C.
The amount of genomic DNA of each isolate was determined by mixing 2 µL of DNA solution with 6 µL of RNA-free deionized water and 2 µL of Tracklt™ Cyan\Orange loading buffer (Invitrogen® Life Technologies Corporation). The samples were loaded into a 1% agarose gel and run at 90 V for 1 h. A low DNA mass ladder (Invitrogen® Life Technologies Corporation) was included to compare and estimate the amount of DNA, which was subsequently diluted to achieve a final concentration of 20 ng µL−1 for polymerase chain reaction (PCR). Concentrations and DNA purity were verified by spectrophotometry using NanoDrop8000® (Thermo Fisher Scientific).
Primers NS1 and ITS4 described by White et al. (Citation1990) were used for the amplification of the internal transcribed spacer (ITS; 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene and internal transcribed spacer 2, complete sequence; and 26S ribosomal RNA gene, partial sequence) gene region, whereas primers ITS3, ITS4 and ITS5 were used for sequencing (Câmara et al. Citation2002). Primers gpd F and gpd R were used for amplification of the partial glyceraldehyde-3-phosphate dehydrogenase gene, and gpd ef, gpd er, gpd if and gpd ir were used for sequencing as described by Câmara et al. (Citation2002). PCR reactions consisted of 2 µL of 10× buffer (MgCl2), 0.25 µL of 10 mM of dNTP mixture, 0.2 µL of 10 pmol of each primer, 0.2 µL of Taq DNA Polymerase (GenScript® USA Inc.), 1 µL of 20 ng of genomic DNA, and 16.15 µL of RNA-free deionized water. PCR was performed on a C1000 Thermal Cycler ® at 94°C for 30 s of denaturation, 57°C for 1 min of annealing, and 72°C for 1.5 min of extension with a final extension at 72°C for 7 min after 35 cycles (Câmara et al. Citation2002; Wang et al. Citation2010).
PCR products were visualized after electrophoresis by illumination under UV light. DNA fragments of approximate sizes of 2.2 kb for ITS and 1.8 kb for gpd were excised and purified with the Bio Basic Inc EZ-10 Spin Column kit. Aliquots of 40–60 ng µL−1 of the purified ITS and gpd amplicons and the respective primers were sent for sequencing to Eurofins Genomics (Eurofins MWG Operon LLC 2016, Louisville, KY).
Sequences of the ITS and the gpd regions were analysed and edited with the DNA Baser Sequence Assembler v4 (Heracle BioSoft SRL Romania, http://www.dnabaser.com). The edited sequences of each target fragment were pooled and a consensus sequence was generated. Sequence data for the ITS and gpd regions were concatenated (Gadagkar et al. Citation2005) for the 33 isolates and seven sequences of species belonging to the Pleosporales retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) (). The partition homogeneity test was carried out using PAUP 4.0a151 to accept or reject the null hypothesis that the two data sets were not significantly different (P > 0.05) to determine whether they could be combined (Swofford Citation2002). Pairwise and multiple alignments were done for each gene separately and for the concatenated data set with the Muscle algorithm. Alignments and the phylogenetic analyses were carried out using Molecular Evolutionary Genetics Analysis Version 6.0 (MEGA6) (Tamura et al. Citation2013).
Table 2. GenBank accession numbers of the partial sequences for the rDNA internal transcribed spacer (ITS; partial sequence of the 18S ribosomal RNA gene, complete sequences of the internal transcribed spacer 1, 5.8S ribosomal RNA gene and internal transcribed spacer 2, partial segment of the 26S ribosomal RNA gene) and glyceraldehyde–3 phosphate dehydrogenase (gpd, partial sequence) gene regions of six Stemphylium spp. and one Alternaria alternata isolate included in the phylogenetic analysis.
The first approach in the phylogenetic analysis was through the Neighbour-Joining (NJ) method (Saitou & Nei Citation1987) as it is considered to be the fastest and most efficient approach (Gadagkar et al. Citation2005). Additionally, the Maximum Likelihood (ML) method was explored to obtain further evidence for the evolutionary relationships among Stemphylium spp.
Branch topologies of phylogenetic trees constructed with the NJ were tested with the bootstrap test with 1000 replications, and the Tamura–Nei model (Tamura & Nei Citation1993) was used to build the highest log likelihood ML phylogenetic tree. The evolutionary distances were calculated with the p-distance method including the 1st, 2nd, 3rd codon positions. Non-coding regions were excluded from the analyses, and gaps and missing data were completely deleted.
The Bayesian inference method using MrBayes 3.1 (Huelsenbeck & Ronquist Citation2001) was used to generate clade credibility support in the ML tree of the concatenated gene regions. This analysis ran for 10 million generations in place of 1 million, trees were sampled every 1000 generations in place of 100 generations; and when the standard deviation of split frequencies was below 0.05 the analysis stopped.
Additionally, estimations of the number of base pair differences per site within and between clusters, of the percentage identity among concatenated sequences of each isolate, and of the number of different nucleotides per site were carried out using pairwise analyses (Tamura et al. Citation2013) and multiple sequence alignment with Muscle logarithm (Clustal2.1 Omega EMBL-EBI, http://www.ebi.ac.uk), and Basic Local Alignment Search Tool logarithm (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi), respectively.
Pathogenicity test of Stemphylium spp. in lentil
A randomized complete block design experiment with four replications was conducted and repeated once to determine the pathogenicity of isolates representing the two clusters of potential stemphylium blight pathogens of lentil of the concatenated ML three in the molecular phylogenetic analysis. For this purpose, isolate SB19 from lentil representing cluster B and SB134 from lentil representing cluster C, were selected, and the Canadian lentil cultivar ‘CDC Robin’ was used as a host. ‘CDC Robin’ is a small-seeded red lentil developed at the Crop Development Centre from ‘CDC Matador’/‘Eston’/‘ESOR – 3 – 6 – 1’ in 1992 and was registered in 1999 (Vandenberg et al. Citation2002). Six seeds were planted in 10 × 10 cm pots filled with Sunshine® mix #4 (Sun Gro Horticulture Canada Ltd, Vancouver, BC) mixed (3:1) with Supreme® perlite (Perlite Supreme Comp., Portland, OR). Four replicate pots per isolate were prepared and after emergence, plants were fertilized with fertilizer solution prepared with PlantProd® 20–20–20 (Master Plant-Prod Inc., Brampton, ON) once a week.
Isolates were grown for 7 d on V8-PDA in 9 mm Petri dishes incubated under light conditions on the bench top at a temperature of approximately 25°C, at which point more than 80% of the plate was covered with mycelia and conidia. Conidia were scraped from the surface of the V8-PDA medium and suspended in distilled water. Conidial concentrations were determined with a Neubauer hemocytometer and adjusted to 1 × 104 conidia mL−1. Before inoculation, two drops of Tween® 20 surfactant per L of suspension were added.
Ten days after planting, plants were thinned from six to four plants per pot and were inoculated with an air brush by spraying approximately 3 mL of conidial suspension per plant which was equivalent to run-off. In addition to the inoculated plants, non-inoculated controls were included. To avoid cross-contamination between isolates and between inoculated and non-inoculated plants, pots were wrapped with plastic foil. Plants were incubated for 48 h in high humidity in a misting chamber, after which they were transferred into a growth chamber at 22°/20°C day/night temperatures and with 16 h photoperiod. Fourteen days after inoculation, the plastic wrap was removed and the percentage disease severity on each plant was recorded. The average disease severity of the four plants per pot was used for statistical analysis using the mixed model procedure of SAS. Replicates and repeats were treated as random effects and isolates as fixed effects. Means were compared by Fisher’s least significant difference (P > 0.05).
Symptomatic tissue samples were collected and incubated on V8-PDA for 7 d at 25°C under continuous light. Re-isolation of the pathogens from the symptomatic tissue was used as confirmation of the pathogenicity of the field isolates tested.
Results
Morphological and morphometric isolate description
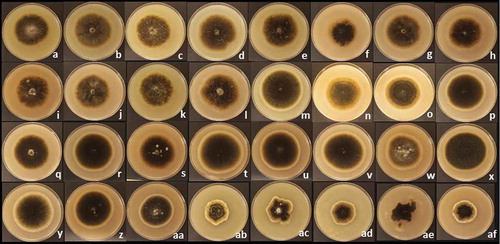
The qualitative description of the 32 isolates after 7 days incubation revealed three distinctive groups sharing colour, texture and shape of colony. The first group consisted of SB11, SB19, SB20, SB40, SB85, SB131, SB133, SB139, SB148, SB149, SB165 and SB167 (). The colonies had a velvety texture and were greyish with slightly concentric rings that turned from dark grey in the centre of the colony to light grey and white towards the edge of the colony. The second group consisted of SB17, SB31, SB49, SB90, SB127, SB134, SB135, SB140, SB141, SB143, SB144, SB146, SB150, SB166 and the ex-type DAOM 195299 (–aa). The colonies also had a velvety texture, but were uniformly grey and had well-defined circular concentric rings that turned to light grey from the centre of the colony to the outer edge. The third group with similar colony features included SB27, SB32, SB44, SB132 and SB151 (ab–af). The colonies were greyish with irregular concentric rings ranging from dark grey in the centre of the colony to light grey and white at the outer edge.
Fig. 1 (Colour online) Colonies of 31 field isolates of Stemphylium spp. and the ex-type of S. botryosum DAOM 195299 after 7 days incubation on V8-PDA medium at 25°C under continuous light. Group 1: (a) SB11, (b) SB19, (c) SB20, (d) SB40, (e) SB85, (f) SB131, (g) SB133, (h) SB139, (i) SB148, (j) SB149, (k) SB165 and (l) SB167; Group 2: (m) SB17, (n) SB31, (o) SB49, (p) SB90, (q) SB127, (r) SB134, (s) SB135, (t) SB140, (u) SB141, (v) SB143, (w) SB144, (x) SB146, (y) SB150, (z) SB166 and (aa) DAOM 195299; Group 3: (ab) SB27, (ac) SB32, (ad) SB44, (ae) SB132 and (af) SB151.
The length and width of conidia of the 32 isolates ranged from 24 to 44 µm and 17 to 28 µm, respectively. Only slight variation in conidial colour, shape and septation were observed among isolates (). Analysis of conidial length and width revealed differences among isolates (P < 0.0001). Simple linear contrasts between the ex-type DAOM 195299 of S. botryosum and field isolates revealed that the length of SB32 conidia was similar to those of the ex-type (P = 0.62), whereas the width of conidia of isolates SB19, SB85, SB127, SB131, SB149, SB151 and SB167 were similar to that of the ex-type (P ≥ 0.06). However, based on Wiltshire’s description (1938), conidia of S. botryosum range from 24 to 39 µm in length and 19 to 31 µm in width, and conidia of other isolates also overlap either in width, length or both with those measurements.
Molecular phylogenetic analysis
Comparison between phylogenetic trees built with the NJ and ML methods revealed similarities in tree topologies for each gene region (Supplemental Figures S1 and S2). The partition homogeneity test for the ITS and gpd gene regions estimated 930 constant characters, 29 variable characters that were parsimony-uninformative and 127 parsimony-informative characters out of 1086 characters through the branch-swapping algorithm (tree-bisection-reconnection). The test resolved that the data sets from the two loci were congruent and were not significantly different from each other, hence the sequences of the ITS and gpd were combined. The phylogenetic trees of the concatenated gene regions using NJ and ML showed similarities in the topology with two groups that were strongly supported with bootstrap values of 99% and 95%, and 95% and 85% for NJ (Supplemental Figure S3) and ML () trees, respectively. The NJ method involved a total of 1086 positions whereas the ML method involved 860 positions from the 40 sequences and is discussed in detail here.
Following Câmara et al. (Citation2002), the two clusters of the phylogenetic tree here were designated as B and C. Both clusters, B and C, from this tree had a credibility clade support of 100% deducted by using the heuristic clade inference (). The average base pair differences per site over all sequences within cluster B was 0.002, which was the same among sequences within cluster C, whereas there was an average of 0.013 base pair differences per site between clusters B and C. A much higher average of 0.057 base pair differences per site was estimated when clusters B and C each were compared with the Alternaria sp. out-group. Comparisons of the sequence of the ex-type DAOM 195299 of S. botryosum as the representative of cluster B with SB146 representing cluster C revealed 96% sequence identity with 100% of the query covered, and with 37 different nucleotides estimated with the BLAST algorithm. On the other hand, the Alternaria out-group revealed 24% and 13% of sequence dissimilarity, and 104 and 95 different nucleotides compared with DAOM 195299 and SB146, respectively.
Fig. 2 Conidia of 31 field isolates of Stemphylium spp. and the ex-type of S. botryosum DAOM 195299 after 14 days incubation on V8-PDA medium at 25°C under continuous light. Photographed at 20× magnification with a ZEN Carl Zeiss camera and measured with ZEISS ZEN Digital Imaging for light microscopy. (a) SB11, (b) SB19, (c) SB20, (d) SB40, (e) SB85, (f) SB131, (g) SB133, (h) SB139, (i) SB148, (j) SB149, (k) SB165 and (l) SB167, (m) SB17, (n) SB31, (o) SB49, (p) SB90, (q) SB127, (r) SB134, (s) SB135, (t) SB140, (u) SB141, (v) SB143, (w) SB144, (x) SB146, (y) SB150, (z) SB166, (aa) DAOM 195299, (ab) SB27, (ac) SB32, (ad) SB44, (ae) SB132 and (af) SB151. Bar = 10 µm.
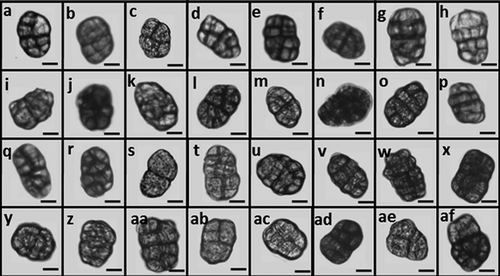
Fig. 3 Maximum likelihood phylogenetic tree (log likelihood = −1930.71) based on the concatenated sequences of the internal transcribed spacer (ITS) and the glyceraldehyde-3-phosphate dehydrogenase (gpd) gene regions for 31 field isolates of Stemphylium spp., the ex-type of S. botryosum DAOM 195299 and an Alternaria sp. sequenced in this study, and the sequences of ex-type/type of six Stemphylium spp. and one isolate of A. alternata retrieved from GenBank. Bootstrap values ≥50% and Bayesian clade support are shown above or below branches. The evolutionary analyses were conducted in MEGA6. Clusters were labelled B and C following Câmara et al. (Citation2002).
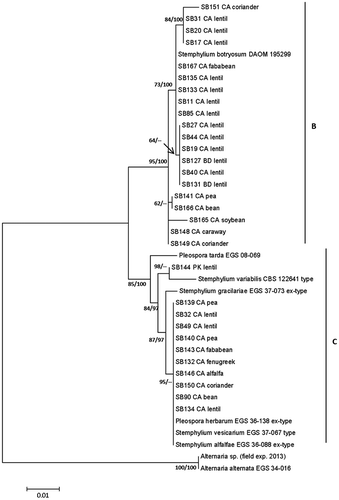
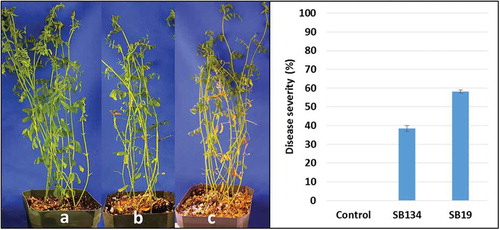
Fig. 4 (Colour online) Stemphylium blight on ‘CDC Robin’ caused by field isolates SB19 and SB134. Left: non-inoculated ‘CDC Robin’ plants (a), ‘CDC Robin’ after inoculation with SB134 (b) and SB19 (c). Right: Stemphylium blight severity (%) on lentil cultivar ‘CDC Robin’ 14 days after inoculation with field isolates SB19 and SB134 at 1 × 104 conidia mL−1.
Eleven of the 14 Canadian field isolates from lentil belong to cluster B whereas three isolates belong to cluster C. In cluster B, isolates SB17, SB20 and SB31 from lentil had two out of 1064 nucleotides different from DAOM 195299, whereas SB151 from coriander had 10 out of 1062 nucleotides different from the ex-type. The sequences of DAOM 195299 were identical with SB167 from faba bean, and with SB11, SB85, SB133 and SB135 from lentil. Lentil isolates SB19, SB27, SB40, SB44, SB127 and SB131 varied in 13 out of 1064 nucleotides from DAOM 195299 whereas isolates SB141 from pea and SB166 from bean varied in 22 out of 1049 nucleotides. Located on a single branch in cluster B, the isolate SB165 from soybean had 16 out of 1063 different nucleotides compared with DAOM 195299. The sequence of DAOM 195299 compared with SB148 from coriander and SB149 from caraway differed in nine out of 1062 different nucleotides.
In cluster C, sequences of 13 isolates including 10 field isolates from a range of host plants and the retrieved sequences of S. vesicarium, S. alfalfae and P. herbarum had identical sequences at the ITS and gpd. The sequences of these 13 isolates differed by six out of 988 from Pleospora tarda EGS 08-073 and by 10 out of 1058 nucleotides from S. gracilariae EGS 37-073, both of which were located on single branches in cluster C. Field isolates SB144 from lentil and S. variabilis CBS 122641 shared a branch in cluster C and had 10 out of 1061 and 19 out of 960 different nucleotides, respectively, compared with the group of 13 identical isolates.
Pathogenicity test
Fourteen days after inoculations, tan-coloured lesions ranging from pinhead-sized to large-sized spots completely covering leaflets were observed on ‘CDC Robin’ plants inoculated with the two isolates. In addition, leaflet drop, one of the characteristic initial symptoms of stemphylium blight, was also observed. No symptoms were observed on non-inoculated control plants. Analysis of variances revealed a significant effect of isolates on disease severity (P < 0.0001). With 58% stemphylium blight severity, isolate SB19 caused significantly more disease than SB134 with 38% on ‘CDC Robin’ (). Both isolates were re-isolated from diseased tissue confirming that they are pathogenic on lentil.
Discussion
Qualitative culture descriptions of isolates of Stemphylium spp. from different hosts and the ex-type DAOM 195299 of S. botryosum revealed some variation among isolates, and three morphological groups based on colony characteristics were distinguished. Incubating at the same temperature used here (25°C), but on PDA, Rahman et al. (Citation2010) observed that several isolates of S. botryosum can develop slight differences in colony features. Similarly, Hosen et al. (Citation2009) described significant variation in colony features among four isolates of S. botryosum obtained from lentil with stemphylium blight symptoms when those isolates were cultured at 25°C on lentil dextrose agar. Both groups of authors described an average colony of S. botryosum as greenish brown to black of velvety texture with a peripheral white ring, which was similar to the description of the colonies of all 31 isolates in this study including DAOM 195299. Only conidia of one isolate (SB32) from Canadian lentil shared the same length, and conidia of another four isolates from lentil (SB19, SB85, SB127 and SB131), two from coriander (SB149 and SB151) and one isolate from faba bean (SB167) shared the width with conidia of DAOM 195299. However, based on Wiltshire’s (Citation1938) size range for S. botryosum, conidia of other isolates also overlapped in size, and any separation based on morphology did not correlate to the grouping based on molecular phylogeny. The limitations of morphological description as a tool for the differentiation among closely related species within the genus Stemphylium have been pointed out previously (Câmara et al. Citation2002; Wang et al. Citation2010), emphasizing the importance of a molecular phylogeny to support species differentiation.
The molecular phylogenetic analyses of ITS and gdp sequences of 31 field isolates presumed to be S. botryosum and six accessions of other Stemphylium spp. accessed from GenBank, irrespective of the approach for analysis, revealed one strongly supported Stemphylium group, similar to the previously described monophyletic group of Stemphylium spp. (Câmara et al. Citation2002; Inderbitzin et al. Citation2009; Wang et al. Citation2010). While the ITS is considered the barcoding region for fungi (Crous et al. Citation2014), gpd is one of at least three additional genes (gpd, EF-1 alpha and vmaA-vpsA) that have been used for evolutionary reconstruction of species in the genus Stemphylium (Inderbitzin et al. Citation2009; Pei et al. Citation2011; Deng et al. Citation2014; Kurose et al. Citation2015; Graf et al. Citation2016). The two methods of phylogenetic analyses used here (NJ, ML) were consistent in separating Stemphylium spp. from the out-group of Alternaria with strong cluster support through bootstrap values of 85–100%. Similar bootstrap values of 100 and 97% for the clusters of Stemphylium–Pleospora and Alternaria alternata, respectively, based on combined ITS and gpd sequences were reported by Wang et al. (Citation2010). This clade support based on bootstrap values was higher than those reported to support the separation of the genera Alternaria, Ulocladium and Stemphylium (73, 82 and 100%, respectively) using ITS sequences only (Chou & Wu Citation2002). Lower bootstrap values of 58–100% for cluster support to separate Alternaria spp. and Setosphaeria spp. from Stemphylium–Pleospora spp. using combined ITS and gpd sequences were reported by Câmara et al. (Citation2002). Tree topology and bootstrap values of 79–100% also supported the separation of the genera Ophiosphaerella and Phaeosphaeria and Pleospora using phylogenetic analyses of the ITS sequences (Câmara et al. Citation2000). Alternaria was used as an out-group since it is a monophyletic genus (Berbee et al. Citation1999) with many morphological similarities, but previously shown to be distinct from Stemphylium (Yanez Citation2001).
Among Stemphylium isolates, two clusters could be discerned based on the topology of each tree, derived from single genes or from the concatenated sequences. Groups from the ML phylogenetic tree were supported with strong bootstrap values of 95 and 85% for clusters B and C, respectively. Similarities among isolates within cluster B ranged from 95 to 100% and within cluster C were from 97 to 100%. These values were similar to, or higher than, values used to separate isolates of Ophiosphaerella agrotis (similarity among isolates from 87 to 100%) from Ophiosphaerella spp. (similarity among isolates from 95 to 97%) by Câmara et al. (Citation2000). However, the similarity values are lower than 99.1% used to separate species of S. solani and S. globuriferum using ITS sequences only (Hanse et al. Citation2015). Furthermore, the base pair differences per site (0.013) between the two clusters was the same as the pair differences per site (0.013) used to separate S. lycopersici from S. vesicarium by Yanez (Citation2001). Therefore, the similarity shared among isolates within each group and the difference in the nucleotide differences between groups may support the assumption that clusters B and C represent two distinct species of Stemphylium.
The location of P. tarda EGS 08-069 in the phylogenetic tree revealed that it is more closely related to S. gracilariae and the species representing isolates in cluster C than to S. botryosum. Indeed, a comparison of sequences by the BLAST algorithm in the GenBank data base revealed that sequences of P. tarda EGS 08-069 and P. herbarum var. herbarum CBS 191.86 were identical (data not shown). Pleospora tarda EGS 08-069 was listed as ex-type/type by Inderbitzin et al. (Citation2009), but although Simmons (Citation1985) examined that isolate, he designated EGS 04-118C originating from Canada and accessioned as DAOM 195299 (QM 1379; ATCC 18522) in the Canadian culture collection as the type. Based on results here, P. tarda EGS 08–069 is not identical to S. botryosum.
Whereas isolates in cluster B can be confirmed as S. botryosum with some confidence based on the comparison with the ex-type DAOM 192599 of S. botryosum, the species identity of isolates in cluster C, including isolates from lentil, pea, faba bean, fenugreek, alfalfa, bean and coriander is not clear since three validly described species of Stemphylium have identical sequences (as retrieved from GenBank) suggesting that they are in fact the same species. Pleospora herbarum, P. alfalfa, P. tomatonis, P. sedicola and S. vesicarium clustered together as one species in the phylogenetic analyses based on four loci (Inderbitzin et al. Citation2009), indicating that they may represent one single species. This confirms that at least two species, S. botryosum and a second species (cluster C) infect lentil in Saskatchewan, considering that pathogenicity testing here of two field isolates representing the two possible Stemphylium species were causing stemphylium blight symptoms on lentil to different degrees.
In conclusion, S. botryosum was confirmed as one of the causal pathogens of stemphylium blight of lentil reported by other authors, but at least one other species is also associated with this disease. The recognition of multiple pathogens associated with stemphylium blight is important for resistance breeding efforts because resistance has to be evaluated in response to all potentially pathogenic species. It is clear from this study as well as previous published research (e.g. Câmara et al. Citation2002; Inderbitzin et al. Citation2009) that an extensive revision of the genus Stemphylium is warranted to clarify the number of species in this genus, so that an accurate identification of the pathogens causing stemphylium blight on a wide range of host species can be accomplished.
TCJP 1378728 Fig S1
Download TIFF Image (122 KB)TCJP 1378728 Fig S2
Download TIFF Image (122.4 KB)TCJP 1378728 Fig S3
Download TIFF Image (114.4 KB)Supplemental data
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2017.1378728
Additional information
Funding
References
- Berbee ML, Pirseyedi M, Hubbard S. 1999. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycology. 91:964–977.
- Câmara MPS, O’Neill NR, Berkum P. 2002. Phylogeny of Stemphylium spp. based on ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycology. 94:660–672.
- Câmara MPS, O’Neill NR, Dernoeden PH, Palm ME. 2000. Ophiosphaerella agrotis sp. nov. and its relationship to other species of Ophiosphaerella. Mycology. 92:317–325.
- Chou H, Wu W. 2002. Phylogenetic analysis of internal transcribed spacer regions of genus Alternaria, and the significance of filament-beaked conidia. Mycol Res. 106:164–169.
- Crous PW, Giraldo A, Hawksworth DL, Robert V, Kirk PM, Guarro J, Groenewald JZ. 2014. The genera of fungi: fixing the application of type species of generic names. IMA Fungus. 5:141–160.
- Deng JX, Paul NC, Li MJ, Cho HS, Lee HB, Yu SH. 2014. Stemphylium platycodontis sp. nov., isolated from Platycodon grandiflorus in Korea. Mycol Prog. 13:477–482.
- Gadagkar SR, Rosenberg MS, Kumar S. 2005. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zool. 304B(1):64–74.
- Graf S, Bohlen-Janssen H, Miessner S, Wichura A, Stammler G. 2016. Differentiation of Stemphylium vesicarium from Stemphylium botryosum as causal agent of the purple spot disease on asparagus in Germany. Eur J Plant Pathol. 144:411–418.
- Hanse B, Raaijmakers EEM, Schoone AHL, Van Oorschot PMS. 2015. Stemphylium sp., the cause of yellow leaf spot disease in sugar beet (Beta vulgaris L.) in the Netherlands. Eur J Plant Path. 142:319–330.
- Hosen MI, Ahmed AU, Zaman J, Ghosh S, Hossain KMK. 2009. Cultural and physiological variation between isolates of Stemphylium botryosum the causal agent of stemphylium blight disease in lentil (Lens culinaris). World J Agric Sci. 5:94–98.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. 2005. Lateral transfer of mating system in Stemphylium. Proc Nat Acad Sc USA 102:11390–11395.
- Inderbitzin P, Mehta YR, Berbee ML. 2009. Pleospora species with Stemphylium anamorphs: a four locus phylogeny resolves new lineages yet does not distinguish among species in the Pleospora herbarum clade. Mycology. 101:329–339.
- Kurose D, Misawa T, Suzui T, Ichikawa K, Kisaki G, Hoang LH, Furuya N, Tsuchiya K, Tsushima S, Sato T. 2015. Taxonomic re-examination of several Japanese Stemphylium strains based on morphological and molecular phylogenetic analyses. J Gen Plant Pathol. 81:358–367.
- Pei Y, Wang Y, Geng Y, O’Neill NR, Zhang X. 2011. Three novel species of Stemphylium from Sinkiang, China: their morphological and molecular characterization. Mycol Prog. 10:163–173.
- QIAGEN. 2012. Sample and assay technologies: quick-start protocol, DNeasy plant mini kit handbook [Internet]. [ cited 2013 May 20]. http://www.quiagen.com/.
- Rahman T, Ahmed AU, Islam MR, Hosen MI. 2010. Physical study and both in vitro and in vivo antifungal activities against Stemphylium botryosum causing stemphylium blight disease in lentil (Lens culinaris). Plant Path J. 9:179–187.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol Evol. 4:406–425.
- Simmons EG. 1967. Typification of Alternaria, Stemphylium, and Ulocladium. Mycology. 59:67–92.
- Simmons EG. 1969. Perfect states of Stemphylium. Mycology. 61:1–26.
- Simmons EG. 1985. Perfect states of Stemphylium II. Sydowia Ann Mycol Ser II. 38:284–293.
- Swofford DL. 2002. PAUP∗. Phylogenetic analysis using parsimony (∗and other methods). Version 4.0a151. Sunderland (MA): Sinauer Associates.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molec Biol Evol. 10:512–526.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molec Biol Evol. 30:2725–2729.
- Vandenberg A, Kiehn FA, Vera C, Gaudiel R, Buchwaldt L, Dueck S, Wahab J, Slinkard AE. 2002. Cultivar description CDC Robin lentil. Can J Plant Sci. 82:111–112.
- Wallroth FG. 1833. Flora cryptogamica Germaniae, pars posterior. Nuremberg: J. L. Schrag; p. 300. https://books.google.ca/books?id=fDw-AAAAcAAJ&ots=nSz7DZmP5g&dq=Flora%20cryptogamica%20Germaniae&lr&pg=PA1#v=onepage&q=Flora%20cryptogamica%20Germaniae&f=false. [Accessed on 7.10.2013]
- Wang Y, Geng Y, Pei Y, Zhang X. 2010. Molecular and morphological description of two new species of Stemphylium from China and France. Mycology. 102:708–717.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and amplifications. New York (NY): Academic Press; p. 315–322.
- Wiltshire SP. 1938. The original and modern conceptions of Stemphylium. Trans British Mycol Soc. 21:211–239.
- Yanez MJM. 2001. Molecular and morphological analyses of Alternaria and Stemphylium from onion [ MSc thesis]. Ann Arbor (MI): Cornell University.
