Abstract
Leaf spots caused by plant pathogenic fungi are among the main diseases affecting orchid plants worldwide. A leaf spot disease was widespread in a nursery of cattleya orchid (Cattleya lueddemanniana var. lueddemanniana) in Lampang Province, Thailand in 2016. A fungus was isolated from leaf lesions and identified as Neoscytalidium orchidacearum based on morphological characteristics and the identification was confirmed using a molecular phylogenetic analysis of the combined internal transcribed spacer and large subunit regions of ribosomal DNA sequences. The isolated fungus caused spot symptoms on inoculated orchid leaves similar to symptoms observed in the field. This is the first report of orchid leaf spot disease caused by N. orchidacearum.
Résumé
Les taches foliaires causées par les champignons pathogènes sont les maladies les plus courantes chez les orchidées, et ce, dans le monde entier. En 2016, une tache foliaire s’était répandue dans une pépinière de cattleya (Cattleya lueddemanniana var. lueddemanniana) de la province de Lampang, en Thaïlande. Un champignon a été isolé à partir de lésions prélevées sur des feuilles et, en se basant sur les caractéristiques morphologiques, a été identifié en tant que Neoscytalidium orchidacearum. Son identité a été confirmée par analyse phylogénétique combinée de l’espaceur transcrit interne et des régions de la grande sous-unité des séquences de l’ADN ribosomique. Le champignon isolé causait des symptômes de taches sur les feuilles inoculées d’orchidées, semblables à ceux observés en plein champ. Il s’agit de la première mention de tache foliaire causée par N. orchidacearum chez l’orchidée.
Introduction
Orchids are a large family of flowering plants that are widely distributed in temperate and tropical regions (Dresslor, Citation1993; Brundrett et al., Citation2001). They are cultivated for their commercial value as flowers and have been successfully hybridized and a range of varieties are available (Brundrett et al., Citation2001; Yam & Arditti, Citation2009). About 185 genera and 1300 species are found in Thailand, where orchid growing started as a hobby about 100 years ago. In 2012, the estimated area of orchid cultivation in Thailand was 3003 ha (Thammasiri, Citation1997). Currently, orchids have the highest value of cut-flower crops in Thailand. Important exported commercial orchid cut-flowers from Thailand are in the genera Ascocentrum, Cattleya, Cymbidium, Dendrobium, Doritis, Paphiopedilum, Phalaenopsis and Vanda (Thaithong, Citation1999; Thammasiri, Citation2015, Citation2016) with an export value of about US$60 million in 2014 (Department of Agricultural Extension, Citation2015). In recent years, as areas planted with orchids have increased, more plantings occur on less suitable sites, and the incidence and severity of diseases have increased.
Damage caused by diseases can result in considerable losses for growers in terms of yield and quality. Many diseases caused by fungi, bacteria and viruses can affect the leaf, stem or root of orchids in both seedling and mature stages (Light, Citation1995; Kawate & Sewake, Citation2014; Han et al., Citation2015). Leaf spot diseases of orchid can be caused by plant pathogenic fungi from several genera, including Alternaria, Cladosporium, Cercospora, Colletotrichum, Fusarium, Guignadia, Phyllosticta, Phytophthora, Pseudocercospora and Septoria (Light, Citation1995; Silva & Pereira, Citation2007; McMillan et al., Citation2008; Kawate & Sewake, Citation2014; Han et al., Citation2015; Sun et al., Citation2017).
Neoscytalidium is a fungal genus within the family Botryosphaeriaceae of the order Botryosphaeriales (Crous et al., Citation2006). Currently, three species of Neoscytalidium have been described: N. dimidiatum, N. novaehollandiae and N. orchidacearum (Crous et al., Citation2006; Phillips et al., Citation2013; Huang et al., Citation2016). Neoscytalidium dimidiatum and N. novaehollandiae have been reported from America, Australia, Brazil, China, Iraq, Israel, Malaysia and Oman as plant pathogens (Farr et al., Citation1989; Elshafie & Ba-Omar, Citation2001; Crous et al., Citation2006; Pavlic et al., Citation2008; Ray et al., Citation2010; Al-Saadoon et al., Citation2012; Ezra et al., Citation2013; Mohd et al., Citation2013; Phillips et al., Citation2013; Machado et al., Citation2014; Yi et al., Citation2015). Neoscytalidium dimidiatum can cause a human skin infection and is associated with rhinosinusitis (Bakhshizadeh et al., Citation2014). The remaining species, N. orchidacearum, has been reported as a saprophytic fungus on dead leaves of orchid (Huang et al., Citation2016).
Three fungal species, Cer. habenariicola, Phy. piriformis and Pseu. dendrobii, have been previously reported as the causal agents of leaf spot diseases on orchid in Thailand (The Organic Support Agriculture Association Thailand, Citation2001–2017; To-Anun et al., Citation2011). A leaf spot disease was observed to be widespread in a nursery of cattleya orchid in Lampang Province, Thailand in 2016. Approximately 65% of the plants were affected, with some of the plants being severely infected. The objective of this study was to identify and describe the causal agent of this disease using morphological and molecular techniques, and pathogenicity tests.
Materials and methods
Sample collection and fungal isolation
Five orchid leaves showing typical symptoms of leaf spot were collected from Cattleya lueddemanniana var. lueddemanniana Rchb. f. plants growing in a nursery in Lampang Province, Thailand during June 2016. Fungi were isolated from the leaf spot lesions following the method described by Han et al. (Citation2015), with some modifications. Small sections (5 × 5 mm) were cut from around necrotic sites and surface-sterilized with 70% ethanol for 1 min, followed by 0.5% NaClO for 1 min, and then rinsed with sterile distilled water three times, before placing on potato dextrose agar (PDA; CONDA®, Spain) supplemented with chloramphenicol (250 ppm; Sigma-Aldrich, Germany). After 5 days of incubation at 25°C, the hyphal tips of emerging colonies were transferred onto new PDA dishes to obtain pure cultures.
Pathogenicity test
Three fungal isolates of different morphology (CMU286, CMU287 and CMU288) obtained from diseased host tissues were used as inocula, which consisted of conidia harvested from 1 week-old cultures on PDA of each isolate. The healthy leaves of commercial cattleya orchid (C. lueddemanniana var. lueddemanniana) were detached and rinsed with tap water, air-dried at room temperature (25 ± 2°C), and the surfaces disinfested with 70% ethanol (Sezer & Dolar, Citation2012). The leaves were then placed in a plastic box with wet, sterilized filter paper. A conidial suspension (20 μL of 5 × 105 conidia mL−1) of each fungal isolate was separately placed onto the intact surface of an orchid leaf according to the procedures described by Sun et al. (Citation2017). Sterile distilled water was used as a control. The plastic boxes were then placed in a growth chamber at 25°C with 12 h of light from fluorescent lamps (1500 lx) and ~70% relative humidity. Ten healthy leaves per isolate and control treatments were inoculated. The symptoms of leaf spot were monitored over 1 week. All treatments were repeated twice with three replicates of each treatment.
Fungal identification
Morphology
Conventional morphological characters tentatively identified fungal isolates collected in this study to genus and species (Ellis, Citation1971; Phillips et al., Citation2013; Huang et al., Citation2016). The morphological data of anatomical features e.g. mycelia, conidiophore and conidia, were based on at least 50 measurements of each structure under a light microscope (Olympus CX51, Japan). Production of fungal conidiomata was induced by cultivation on 2% water agar with sterilized pine needles and incubated under near-UV light at 25°C following the method described by Crous et al. (Citation2006).
Molecular
Molecular methods were used to confirm the identification of the fungal isolate. Fungal isolate CMU287 was grown on PDA at 25 ± 2°C in darkness for 1 week and genomic DNA was extracted from mycelium using a DNA Extraction Mini Kit (FAVORGEN, Taiwan) following the manufacturer’s protocol. The internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) was amplified by polymerase chain reaction (PCR) using ITS4 and ITS5 primers (White et al., Citation1990), under the following thermal conditions: 95°C for 2 min, and then 30 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 1 min, followed by a final extension at 72°C for 10 min using a peqSTAR 2X Thermocycler (PEQLAB, UK). The large subunit (LSU) of rDNA was amplified with LROR and LR5 primers (Vilgalys & Hester, Citation1990) under the following thermal conditions: 94°C for 2 min, and then 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. PCR products were purified using NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel, Germany). The purified PCR products were sequenced by 1ST BASE Company (Kembangan, Malaysia). Sequences were assembled and edited with Sequencher 4.0 (Gene Codes Corp., USA), and used to query GenBank via BLAST (http://blast.ddbj.nig.ac.jp/top-e.html).
Phylogenetic analysis
For phylogenetic analysis, sequences from this study and those of nine isolates of Neoscytalidium spp. obtained from the GenBank database were used. The multiple sequence alignment of the combined ITS and LSU sequences was conducted using MUSCLE (Edgar, Citation2004) and the alignment was deposited in TreeBASE under the number 21090. A maximum likelihood (ML) phylogenetic tree was constructed using RAxML v7.0.3 (Stamatakis, Citation2006), applying the rapid bootstrapping algorithm for 1000 replications using the GTRGAMMA model. Bayesian phylogenetic analyses were carried out using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes version 3.2 (Ronquist et al., Citation2012). Botryosphaeria dothidea and Cophinforma atrovirens sequences were used as outgroups.
Results and discussion
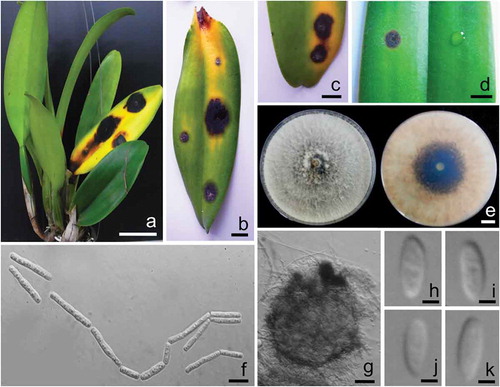
The initial symptoms of the disease on orchid in the field appeared as dark spots that gradually increased from 5 to 20 mm in diameter, changing from circular to elliptical lesions. Leaf spots were dark brown to black, surrounded by a yellowish to rust brown zone within 2 weeks (–). Three fungal isolates, CMU286, CMU287 and CMU288, were obtained from diseased host tissues.
Fig. 1 (Colour online) a, b, c, Field symptoms of leaf spot disease on cattleya orchid (Cattleya lueddemanniana var. lueddemanniana) caused by Neoscytalidium orchidacearum. d, Leaf spot disease of orchid inoculated with N. orchidacearum CMU287 (left) and control (right). e, Colonies of N. orchidacearum CMU287 from the top (left) and bottom (right) on PDA at 25°C after 1 week. f, Arthric chains of conidia growing on PDA. g, Conidiomata formed on pine needles in culture. h, i, j, k, Conidia of coelomycetous state. Scale bars: A = 50 mm; b, c, d and e = 10 mm; f = 10 µm; g = 50 µm; h, i, j and k = 3 µm.
Pathogenicity test
The pathogenicity test indicated that disease symptoms similar to those seen in the field were observed on the leaf inoculated with a conidial suspension of isolate CMU287 (), while no disease symptoms were observed in the control leaf or those inoculated with conidial suspension of isolates CMU286 and CMU288 after 1 week of incubation. Fungal isolates recovered from inoculated diseased lesions had the same morphological features as isolate CMU287, thereby fulfilling Koch’s postulates.
A pure culture of isolate CMU287 was deposited in the Sustainable Development of Biological Resources Laboratory (SDBR), Faculty of Science, Chiang Mai University, Thailand as number SDBR-CMUEN287.
Morphological characteristics
The colonies of isolate CMU287 on PDA grew to 70–75 mm in diameter in 3 days at 25°C. The colonies were flat, downy to woolly, with a white to cream surface, becoming greenish olivaceous from the middle within 1 week. The reverse side was initially white to light orange and later dark grey to brown (). Conidia occurred in arthric chains in aerial mycelium and were 4–30 × 1.5–5 µm (mean 17 × 3.75 µm, n = 50), cylindrical, oblong-obtuse to doliiform, truncate at the ends, hyaline becoming light brown, and 0–2 septate (). These morphological characteristics assigned this fungus to the genus Neoscytalidium (Crous et al., Citation2006; Phillips et al., Citation2013). Conidiomata were observed after 2 weeks of induction. Conidiomata were 200–450 µm in diameter, globose to subglobose, brown to dark brown with 2–3 wall layers which were with 25–70 µm thick, and a short neck with a central ostiole (). Conidiophores were reduced to conidiogenous cells. Conidiogenous cells were hyaline, smooth, cylindrical to subcylindrical and 5–15 × 1.5–3 µm (mean 5.6 × 2.25 µm, n = 50). Conidia were hyaline, smooth, ellipsoidal to oval, aseptate becoming 2–3-septate, and 10–15 × 3–5 µm (mean 12.5 × 2.5 µm, n = 50) (–k). Based on these characteristics, the fungus was identified as N. orchidacearum (Huang et al., Citation2016). Two types of conidia in conidiomata of N. novaehollandiae clearly separate it from N. dimidiatum and N. orchidacearum (Crous et al., Citation2006; Phillips et al., Citation2013; Huang et al., Citation2016). Neoscytalidium orchidacearum is distinguished from N. dimidiatum by its hyaline conidia (Phillips et al., Citation2013; Bakhshizadeh et al., Citation2014; Huang et al., Citation2016).
Molecular identification
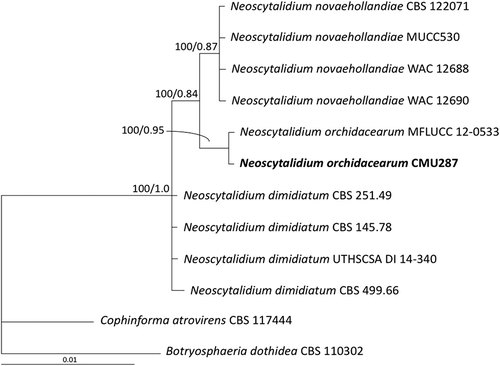
The ITS and LSU sequences of isolate CMU287 were deposited in the GenBank database as accession numbers KY933091 and KY933092, respectively. In the combined ITS and LSU phylogram (), isolate CMU278 clustered with N. orchidacearum MFLUCC 12–0533, with 99 and 100% similarity in ITS and LSU sequences, respectively. Neoscytalidium orchidacearum formed a sister taxon to N. novaehollandiae. Thus, isolate CMU287, obtained from an orchid leaf spot lesion, was identified as N. orchidacearum, based on both morphological and molecular characteristics.
Fig. 2 Phylogram derived from maximum likelihood analysis of combined internal transcribed spacer and large subunit of rDNA sequence data for 12 fungal strains including isolate CMU287 isolated from diseased orchid leaf tissue in this study. The sequences for the other 11 fungal strains were obtained from the GenBank database. Botryosphaeria dothidea and Cophinforma atrovirens were used as outgroups. The numbers above branches represent maximum likelihood bootstrap percentages (left) and Bayesian posterior probabilities (right). Only bootstrap values ≥ 50% are shown. Isolate CMU287 sequenced in this study is indicated in bold.
The leaf spot disease symptom on cattleya orchid caused by N. orchidacearum is similar to those caused by other leaf spot pathogens e.g. Cla. cladosporiodes, F. subglutinans, Phy. capitalensis and Pseu. dendrobii (McMillan et al., Citation2008; Kawate & Sewake, Citation2014; Han et al., Citation2015; Sun et al., Citation2017). There have been no previous reports of N. orchidacearum as a plant pathogen. Therefore, we propose that N. orchidacearum be added as one of the causal agents of leaf spot on cattleya orchid. The source of inoculum of this pathogen is unknown but could originate from dead or decaying leaves in the greenhouse.
Acknowledgements
We are grateful to Dr Eric H.C. McKenzie for improving the English text.
Additional information
Funding
References
- Al-Saadoon AH, Ameen MKM, Hameed MA, Al-Badran A, Ali Z. 2012. First report of grapevine dieback caused by Lasiodiplodia theobromae and Neoscytalidium dimidiatum in Basrah, Southern Iraq. Afr J Biotechnol. 11:16165–16171.
- Bakhshizadeh M, Hashemian HR, Najafzadeh MJ, Dolatabadi S, Zarrinfar H. 2014. First report of rhinosinusitis caused by Neoscytalidium dimidiatum in Iran. J Med Microbiol. 63:1017–1019.
- Brundrett M, Sivasithamparam K, Ramsay M, Krauss S, Taylor R, Bunn E, Hicks A, Karim NA, Debeljak N, Mursidawati S, et al. 2001. Orchid conservation techniques manual, first international orchid conservation congress-training course. Perth: Plant Science, King Park and Botanic Garden.
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Philips AJL, Alves A, Burgess T, Barber P, Groenewald JZ. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Stud Mycol. 55:235−253.
- Department of Agricultural Extension. 2015. Statistics of cut-flowers and orchid production. Bangkok (Thailand): Ministry of Agriculture and Cooperatives.
- Dresslor RL. 1993. Phylogeny and classification of the orchid family. Cambridge: Cambridge University Press.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792−1797.
- Ellis MB. 1971. Dematiaceous Hyphomycetes. Surrey: Commonwealth Mycological Institute.
- Elshafie AE, Ba-Omar T. 2001. First report of Albizia lebbeck dieback caused by Scytalidium dimidiatum in Oman. Mycopathologia. 154:37−40.
- Ezra D, Liarzi O, Gat T, Hershcovich M, Dudai M. 2013. First report of internal black rot caused by Neoscytalidium dimidiatum on Hylocereus undatus (Pitahaya) fruit in Israel. Plant Dis. 97:1513.
- Farr DF, Bills GF, Chamuris GP, Rossman AY. 1989. Fungi on plants and plant products in the United States. Minnesota: APS Press.
- Han KS, Park JH, Back CG, Park MJ. 2015. First report of Fusarium subglutinans causing leaf spot disease on Cymbidium orchids in Korea. Mycobiology. 43:343−346.
- Huang SK, Tangthirasunun N, Phillips AJL, Dai DQ, Wanasinghe DN, Wen TC, Bahkali AH, Hyde KD, Kang JC. 2016. Morphology and phylogeny of Neoscytalidium orchidacearum sp. nov. (Botryosphaeriaceae). Mycology. 44:79−84.
- Kawate M, Sewake KT. 2014. Pest management strategic plan for potted orchid production in Hawaii. In: Tarutani C, editor. Summary of a workshop held on September 30, 2010. Mānoa: University of Hawaii.
- Light MHS. 1995. Growing orchids in the Caribbean. London: Macmillan Education.
- Machado AR, Pinho DB, de Oliveira SAS, Pereira OL. 2014. New occurrence of Botryoshaeriaceae causing black root rot of cassava in Brazil. Trop Plant Pathol. 9:464−470.
- McMillan RT, Palmateer AJ, Vendrame WA. 2008. Cercospora leaf spot caused by Cercospora dendrobii on Dendrobium antennatoum Lindl. and its control. Proc Fla State Hort Soc. 212:335−355.
- Mohd MH, Salleh B, Zakaria L. 2013. Identification and molecular characterizations of Neoscytalidium dimidiatum causing stem canker of red-fleshed dragon fruit (Hylocereus polyrhizus) in Malaysia. J Phytopathol. 161:841–849.
- The Organic Support Agriculture Association Thailand. 2001–2017. Orchid disease compile. [ accessed 2017 Oct 25]. http://www.kokomax.com/product/.
- Pavlic D, Wingfield MJ, Barber P, Slippers B, Hardy GE, Burgess TI. 2008. Seven new species of the Botryosphaeriaceae from baobab and other native trees in Western Australia. Mycologia. 100:851−866.
- Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW. 2013. The Botryosphaeriaceae: genera and species known from culture. Stud Mycol. 76:51−167.
- Ray JD, Burgess T, Lanoiselet VM. 2010. First record of Neoscytalidium dimidiatum and N. novaehollandiae on Mangifera indica and N. dimidiatum on Ficus carica in Australia. Australas Plant Dis Notes. 5:48−50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Sezer A, Dolar FS. 2012. Collectotrichum acutatum, a new pathogen of hazelnut. J Phytopathol. 160:428−430.
- Silva M, Pereira OL. 2007. First report of Guignardia endophyllicola leaf blight on Cymbidium (Orchidaceae) in Brazil. Australas Plant Dis Notes. 2:31−32.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Sun C, Wang T, Shen X, Wang G, Gao Q, Lou B, Shao Y. 2017. First report of leaf spot caused by Cladosporium cladosporioides on Dendrobium officinale in China. Plant Dis. 101:1055.
- Thaithong O. 1999. Orchids of Thailand. Bangkok: Office of Environmental Policy and Planning.
- Thammasiri K. 1997. Orchids in Thailand: a success story. New Delhi (India): Angkor Publishers Ltd.
- Thammasiri K. 2015. Current status of orchid production in Thailand. Acta Horticult. 1078:25–33.
- Thammasiri K. 2016. Thai orchid genetic resources and their improvement. Horticulture. 9:1–13.
- To-Anun C, Hidayat I, Meeboon J. 2011. Genus Cercospora in Thailand: taxonomy and phylogeny (with a dichotomous key to species). Plant Pathol Quar J Fungal Biolog. 1:11–87.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172:4239−4246.
- White TJ, Burns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols, a guide to methods and applications. San Diego: Academic Press; p. 315–322.
- Yam TW, Arditti J. 2009. History of orchid propagation: a mirror of the history of biotechnology. J Plant Biotechnol Rep. 3:1–56.
- Yi RH, Lin QL, Mo JJ, Wu FF, Chen J. 2015. Fruit internal brown rot caused by Neoscytalidium dimidiatum on pitahaya in Guangdong province, China. Australas Plant Dis Notes. 10:13.
