Abstract
Leaf spot diseases occur widely and pose serious impacts on peach production in China. In this study, 60 symptomatic leaf samples were collected from seven provinces in China. Diseased samples were plated on nutrient agar and sucrose-peptone-agar media and purified colonies were identified as Xanthomonas arboricola pv. pruni (Xap) based on morphological features, molecular identification and pathogenicity test. Hypersensitive response and pathogenicity tests were conducted on inoculated tobacco and peach seedlings. Phylogenetic analysis of 16S rRNA gene sequences indicated that the Xap isolates clustered together with Korean, Japan and Taiwan isolates. This is the first report of bacterial leaf spot caused by Xap on peach in China.
Résumé
Les taches foliaires sont des maladies très répandues qui occasionnent de graves problèmes quant à la production de pêches en Chine. Dans cette étude, 60 échantillons de feuilles symptomatiques ont été collectés dans sept provinces. Les échantillons infectés ont été déposés sur un milieu de croissance gélosé et un milieu gélosé au sucrose-peptone. En se basant sur les traits morphologiques, l’identification moléculaire et la pathogénicité, les colonies purifiées ont été identifiées en tant que Xanthomonas arboricola pv. pruni (Xap). Des tests d’hypersensibilité et de pathogénicité sur semis inoculés de tabac et de pêchers ont été menés. L’analyse phylogénétique des séquences du gène 16S de l’ARNr a indiqué que les isolats de Xap appartenaient au meme groupe que les isolats coréens, japonais et taiwanais. Il s’agit de la première mention de la tache foliaire bactérienne causée par Xap chez le pêcher en Chine.
Introduction
Peach (Prunus persica L.) is one of the major stone fruit crops cultivated worldwide. China is the world’s leading producer of peaches both in terms of production area and yield. In China, peach trees have been widely grown in almost all provinces (Yang et al. Citation2011; Li Citation2013). Bacterial leaf spot, caused by Xanthomonas arboricola pv. pruni [Smith] (Xap) is an economically important disease of peach worldwide, and results in significant economic losses on most of the stone fruit trees (Stefani Citation2010; Janse Citation2012). Disease symptoms include lesions on leaves, twigs, fruits and stem cankers (EPPO Citation2016). Moreover, the infected fruit trees show severe defoliation which significantly reduces the yield and quality. In general, fruits affected by this particular disease are unmarketable (Kawaguchi Citation2014). Xap was first reported from the USA on Japanese plum (Smith Citation1903). It is widely disseminated in different parts of the world, causing symptoms on leaves, twigs and fruits (Ritchie Citation1999; EPPO Citation2006, Citation2013, Citation2016). More recently, Xap was reported on apricot (Prunus armeniaca L.), cherry laurel (Prunus laurocerasus L.) and Japanese plum (Prunus salicina Lindl) in Hungary, Taiwan and the Netherlands, respectively (Tjou-Tam-Sin et al. Citation2012; Shen et al. Citation2013; Schwarczinger et al. Citation2017).
To date, bacterial leaf spot disease is only regulated in European and Mediterranean regions using EPPO standards (EPPO Citation2006). This is based on bacterial isolation on growth media, hypersensitive response, pathogenicity, physiological and biochemical tests for isolation of pathogen and identification of Xap from Prunus species. As far as we know, there have been no previous reports on bacterial leaf spot disease caused by Xap on peach cultivars in China. Moreover, the molecular characteristics of Xap have not yet been fully studied in China.
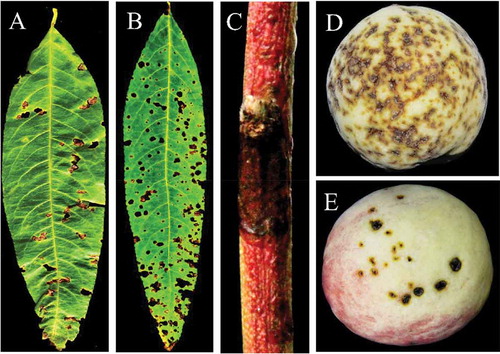
Perforated or shot-hole symptoms were observed on peach leaves in China between 2016 and 2017 (). The symptoms were also observed on twigs () and fruits (). The objective of this study was to isolate and identify the causal agent associated with leaf spot of peach cultivars in China. Xap was detected and bacterial isolates were identified from 27 peach cultivars and 60 leaf samples collected from different peach growing provinces in China using morphological characteristics, molecular identification and pathogenicity tests.
Fig. 1 (Colour online) Distribution of bacterial leaf spot caused by Xanthomonas arboricola pv. pruni in China. The shaded areas are where peach samples were collected in this study.
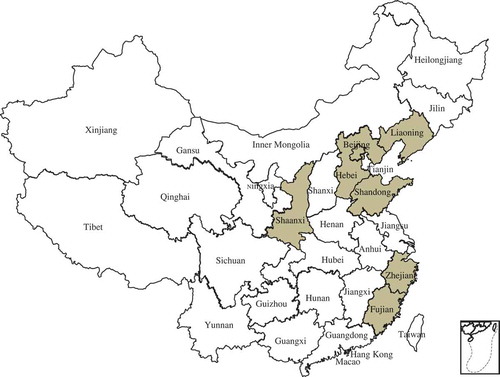
Fig. 2 (Colour online) Typical symptoms of bacterial leaf spot disease on naturally infected peach leaves, twigs and fruits. (a) brown and perforated symptomatic peach leaf collected from Beijing Pinggu; (b) severe symptoms on the entire peach leaf collected from Hebei Changli; (c) dark canker symptomatic peach twig collected from Beijing Pinggu and (d, e) severe brown, circular, water soaked spots and cracking symptoms observed on peach fruit collected from Beijing Pinggu district.
Materials and methods
Sample collection and isolation of the pathogen
Sixty leaf samples of 27 different peach cultivars (‘Beijing 14 and 15‘, ‘Wang 24‘, ‘Zaoyu 9034‘, ‘Huayu’, ‘Zaohongzhu’, ‘Xiahui’, ‘Chiyue’, ‘Yulu’, ‘Hujingmilu’, ‘Wanbaifeng’, ‘Hongyu’, ‘Yufei’, ‘Chaohong’, ‘Zhongyou 4‘, ‘Jinhui’, ‘Liuyefeicheng’, ‘Zhongyou 7‘, ‘Zhongyou 8‘, ‘Mishuo’,‘Dajiubao’, ‘Zaoshuhongburuan’, ‘Dajiubao’, ‘Jixiang’ and ‘Meixiang’) showing perforated leaf symptoms were collected from seven locations () in China between 2016 and 2017. Briefly, peach leaves from the diseased and healthy tissues were cut into 2–3 mm pieces and disinfected with 70% ethanol and then rinsed once with sterile distilled water for 3 min. Suspensions (10–2) were prepared and 10 µL from each sample was spread onto plates containing nutrient agar (NA) and Sucrose-Peptone-Agar (SPA) medium. The plates were incubated for 2–3 days at 28°C. Thereafter, a single colony was taken and placed on NA and SPA for 2 days to obtain pure colonies for the hypersensitive response, molecular identification and pathogenicity tests. All Xap-like isolates were stored at −80°C in NA liquid medium with 25% of glycerol.
Molecular identification
DNA was extracted from 27 isolates using the CTAB method (Doyle Citation1990) and the bacterial DNA extraction kits (Tiangen, Beijing, China) according to the manufacturer’s instructions. The extracted genomic DNA was amplified using primer pair XapY17-F (GACGTGGTGATCAGCGAGTCATTC) and XapY17-R (GACGTGGTGATGATGATCTGC) (Pagani Citation2004) for the ABC transporter ATP-binding protein. PCR assay was performed in a 25 µL reaction mixture containing 2 µL of total DNA, 0.5 µL of each primer (20 µM), 12.5 µL of 2×Taq PCR MasterMix (0.1 U Taq Polymerase µL−1, 500 µM dNTP each, 20 mM Tris-HCL (pH 8.3), 100 Mm KCl, 3 mM MgCl2) (Tiangen, Beijing, China) and 9.5 µL of double distilled water. The PCR was carried out at 94°C for 4 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 60 s and a final extension of 72°C for 10 min. This produced a 943 bp fragment which was cloned and sequenced.
The 16S ribosomal RNA regions of the isolates were amplified using universal primer pair 27-F (AGAGTTTGATCCTGGCTCAG) and 1492-R (TACGGCTACCTTGTTACGACTT) (Lane Citation1991). PCR assay was performed in a 50 µL reaction mixture containing 25 µL of 2×Pfu PCR MaterMix (0.1 U Pfu Polymerase µL−1, 500 µM dNTP each, 50 mM Tris-HCL (pH 8.7), 20 mM KCl, 4 mM MgCl2) (Tiangen, Beijing, China), 2 µL of each primer (10 µM), 4 µL of bacterial genomic DNA as the template and 17 µL of double distilled water. The PCR conditions were 95°C for 5 min followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 2 min and a final extension of 72°C for 10 min.
PCR products were separated in 1.5% agarose gels containing 1×TAE buffer, stained with Gel Red dye (Biotium, Fremont, CA, USA) visualized under UV light, excised and purified using nucleic acid purification kits (Axygen). The purified DNAs were cloned into a pMD18-T vector (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The ligation reaction was used to transform the competent Escherichia coli DHA 5α cells. The positive clones were confirmed by colony PCR from each sample, and two positive clones were sequenced. The nucleotide sequence of the inserted DNA fragment was determined by Sangon (Beijing) using the standard primer M13. The sequenced nucleotide was assembled using DNAMAN software, whereas the sequence alignments were done using CLUSTAL-W. To compare the similarities or homologies, the nucleotide sequences were compared with the sequences available in the GenBank databases using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic analysis of 16S rRNA sequences were conducted using the neighbour-joining method by MEGA6, using the Kimura 2-parameter model method with a bootstrap of 1000 replicates (Tamura et al. Citation2013). Pseudomonas syringae pv. syringae (accession no. DQ318866.1) was used as the out-group taxon.
Hypersensitivity response (HR) and pathogenicity tests
Two tobacco (Nicotiana benthamiana Domin) plants were inoculated with 108 CFU mL−1 of Xap-like suspensions of each isolate from a 3-day old culture grown on NA and SPA at 28°C and grown in a greenhouseat 28°C (day) for 16 h and 25°C (night) for 8 h to assess their hypersensitive response within 24 h. In addition, pathogenicity tests were performed (EPPO Citation2006). Two peach seedlings cv. ‘Qingzhou Mi Tao’ were spray-inoculated with 108 CFU mL−1 of bacterial suspensions of each of 27 isolates from a 3-day old culture grown on NA and SPA at 28°C. The spray-inoculated peach seedlings were maintained in the greenhouse at 28°C (day) for 16 h and 25°C (night) for 8 h. Non-inoculated or distilled water spray-inoculated peach seedlings were used as a negative control. All the spray-inoculated peach seedlings were monitored every 2 days up to 4 weeks for disease development. Bacterial leaf spot symptoms on infected peach leaf tissues were used for isolation on NA and SPA and morphological characteristics and molecular identification of colonies were compared with the original isolates.
Results and discussion
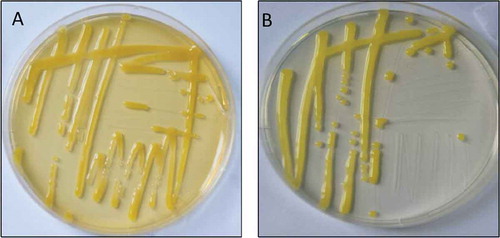
Occurrence of peach leaf spot disease in China
The survey for peach leaf spot diseases was carried out in major peach growing areas (). It was observed that bacterial leaf spot occurred in almost all surveyed peach orchards. The symptoms on peach were similar to those of typical bacterial leaf spot disease of stone fruits (EPPO Citation2006), including leaf spots with severe necrotic brown shot-hole or perforated appearance; severe brown and light green haloes, circular and water-soaked spots and later on necrosis symptoms on peach fruit; and dark spots on twigs (). Twenty-seven isolates of bacteria were obtained from 60 symptomatic peach leaf samples. After 2–3 days of incubation at 28°C on NA and SPA media, all isolates developed bright yellow round colonies which were mucoid (). The recovered colonies were similar to previously reported morphological characteristics of Xanthomonas (Tjou-Tam-Sin et al. Citation2012). Colonies on NA were more bright yellow compared with SPA (). Thus, both media might be useful in future studies for isolation of Xanthomonas from Prunus plant species.
Identification and phylogenetic analysis of 16S ribosomal RNA sequences
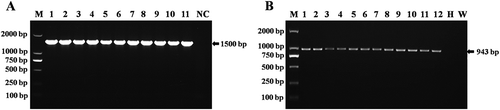
Sequencing of 16S rRNA was performed to identify the causal agent of peach leaf spot disease. The amplified fragment was about 1500 bp in size (). BLASTn search revealed that the highest nucleotide similarity (100%) was with Xap strains (80 476, 80 478, and 80 481) 16S rRNA gene, partial sequence isolated from Taiwan (accession no. KJ156333-35), Xap strain NCPPB416 16S rRNA gene, partial sequence (99%) isolated from Japan (accession no. AB558556) and Xap strain LMG852 16S rRNA gene, partial sequence (99%) isolated from Korea (accession no. FJ606765). The sequences of 16S rRNA region for seven isolates from this study were deposited in the NCBI GenBank (accession nos MF351917–MF351923).
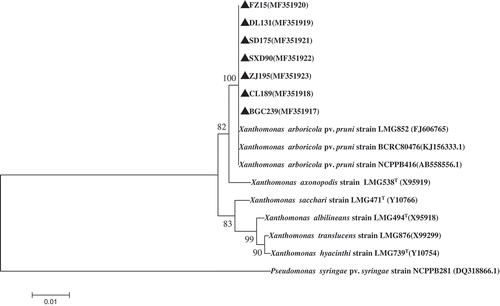
In the phylogenetic tree, all Xap isolates were grouped in the same clade with reference isolates of Xap from Korea (FJ606765), Japan (AB558556) and Taiwan (KJ156333) (). These results indicated that Xap isolates might be the causal agent of leaf spot disease on peach.
Detection of Xap by PCR
The expected size of 943 bp fragment from all 27 isolates was amplified (). The results showed that all 27 peach cultivars were positive for Xap. No signal was obtained from water control and healthy peach seedlings (peach grown in greenhouse) (). This showed that Xap was found in all surveyed provinces and peach cultivars. BLASTn searches indicated that our isolates shared the highest nucleotide identity (99%) with Xap strain CFBP 5530 and XAP HU1 ABC transporter ATP-binding protein (ftsX) isolated from Italy (accession no. HQ896469) and Hungary (accession no. KY039173), respectively. The nucleotide sequence of one isolate (BJ15) was deposited in GenBank (accession no. MF362224).
Fig. 4 (a) PCR amplification fragments of 16S rRNA region from isolates of Xanthomonas arboricola pv. pruni. M: DL2000 DNA marker (TIANGEN, Beijing, China). Lane NC: Negative control. Lanes 1–11: isolates of Xanthomonas arboricola pv. pruni. (b) Detection results of Xanthomonas arboricola pv. pruni by PCR using primer pairs of XapY17-F and XapY-R from naturally infected and greenhouse inoculated peach samples and 3-day old pure culture media. Lane M: DL2000 DNA marker (Tiangen, Beijing, China). Lanes 1–6: isolates of Xanthomonas arboricola pv. pruni from field grown peach samples. W: water control: H: DNA from healthy peach samples. Lanes 7–12: isolates of Xanthomonas arboricola pv. pruni from greenhouse inoculated peach seedlings.
Hypersensitive response and pathogenicity tests
The hypersensitive response (HR) on tobacco leaves resulted in a loss of turgidity, brown colour at the site of inoculation and necrotic symptoms within 24 h after inoculation, but not on water controls (). Pathogenicity tests on leaves of peach seedling were conducted for all 27 isolates and showed typical symptoms of Xap on all inoculated peach seedlings within 10 days following inoculation (EPPO Citation2006). The negative control (distilled water inoculation) did not develop any symptoms. However, the spray-inoculated peach seedlings developed necrotic brown spots which expanded to produce a shot-hole (). The diseased peach leaves were used to re-isolate bacterial strains and confirm the morphological characteristics and molecular identification with the original isolates.
Fig. 5 (Colour online) (a) Hypersensitive response (HR) by Xap isolates on tobacco leaf (Nicotiana benthamiana). (b) Xap isolates inoculated and their symptoms 10 days after inoculation on peach seedlings.
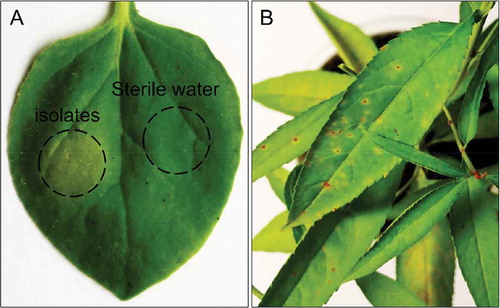
Fig. 6 Phylogenetic tree constructed with the 16S rRNA region sequence of Xanthomonas arboricola pv. pruni isolated in this study (MF351917–MF351923) and other species of Xanthomonads retrieved from GenBank. The tree was constructed by neighbour-joining method from the alignments of 16S rRNA sequences from this study and representative Xanthomonas species from NCBI using MEGA software (version 6) using 1000 bootstrap replicates; only bootstrap values greater than 50% are indicated . Pseudomonas syringae pv. syringae (accession no. DQ318866.1) was used as the out-group taxon.
Generally, bacterial leaf spot disease caused by Xap has been continuously threatening the production and productivity of stone fruit trees around the world (Stefani Citation2010). In recent years, infection by Xap on stone fruit trees has been reported in many different countries (Jami et al. Citation2005; Palacio-Bielsa et al. Citation2010; Pothier et al. Citation2010; Marchi et al. Citation2011; Tjou-Tam-Sin et al. Citation2012; Shen et al. Citation2013; Giovanardi et al. Citation2017; Schwarczinger et al. Citation2017). Peach fruits affected by Xap are unmarketable with lower yield and quality of peach fruits (Janse Citation2012). Severe disease outbreak has been reported in different countries when highly susceptible peach varieties are cultivated (Ritchie Citation1995; Roselló et al. Citation2012; Lamichhane Citation2014). To our knowledge, this is the first report of bacterial leaf spot disease caused by Xap on peach in China. During our orchard surveys, Xap was detected and identified in all peach cultivars and surveyed locations. However, the possible origin of the initial inoculum, susceptibility and resistance of Chinese peach cultivars to Xap infection, disease epidemiology and different control measures of Xap in China need to be studied in detail in future research. These will help design efficient and effective control methods for Xap in China.
Additional information
Funding
References
- Doyle DJ. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- [EPPO] European and Mediterranean Plant Protection Organization. 2006. EPPO standards PM7/64. (1): Diagnostics of Xanthomonas arboricola pv.pruni. EPPO Bulletin. 36:129–133.
- [EPPO] European and Mediterranean Plant Protection Organization. 2013. Xanthomonas arboricola pv.pruni (XANTPR). [Accessed 14 August 2017]. EPPO Global Database. http://gd.eppo.int/taxon/XANTPR/distribution.
- [EPPO] European and Mediterranean Plant Protection Organization. 2016. Xanthomonas arboricola pv.pruni (XANTPR). [Accessed 14 August 2017]. EPPO Global Database. http://gd.eppo.int.
- Giovanardi D, Dallai D, Stefani E. 2017. Population features of Xanthomonas arboricola pv. pruni from Prunus spp. orchards in northern Italy. Eur J Plant Pathol. 147:761–771.
- Jami F, Kazempour MN, Elahinia SA, Khodakaramian G. 2005. First report of Xanthomonas arboricola pv.pruni on stone fruit trees from Iran. J Phytopathol. 153:371–372.
- Janse JD. 2012. Bacterial diseases that may or do emerge, with (possible) economic damage for Europe and the Mediterranean basin: Notes on epidemiology, risks, prevention and management on first occurrence. J Plant Pathol. 94:S4.5–S4.29.
- Kawaguchi A. 2014. Genetic diversity of Xanthomonas arboricola pv. pruni strains in Japan revealed by DNA fingerprinting. J Gen Plant Pathol. 80:366–369.
- Lamichhane JR. 2014. Xanthomonas arboricola disease of stone fruit, almond, and walnut trees: Progress toward understanding and management. Plant Dis. 98:1600–1610.
- Lane DJ. 1991. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York: Wiley; p. 115–175.
- Li SH. 2013. Peach tree. China Agriculture Press; p. 7–8. Beijing, China. (in Chinese).
- Marchi G, Cinelli T, Surico G. 2011. Bacterial leaf spot caused by the quarantine pathogen Xanthomonas arboricola pv. pruni on cherry laurel in central Italy. Plant Dis. 95:74.
- Pagani MC. 2004. An ABC transporter protein and molecular diagnosis of Xanthomonas arboricola pv.pruni causing bacterial spot of stone fruits [ PhD dissertation]. Raleigh: North Carolina State University.
- Palacio-Bielsa A, Roselló M, Cambra MA, Lopez MM. 2010. First report on almond in Europe of bacterial spot disease of stone fruits caused by Xanthomonas arboricola pv. pruni. Plant Dis. 94:786.
- Pothier JF, Pelludat C, Bounter M, Genini M, Vogelsanger J, Duffy B. 2010. First report of the quarantine pathogen Xanthomonas arboricola pv. pruni on apricot and plum in Switzerland. Plant Pathol. 59:404.
- Ritchie DF. 1995. Bacterial spot. In: Ogawa JM, Zehr EI, Bird GW, editors. Compendium of stone fruit diseases. St. Paul: The American Phytopathological Society Press; p. 50–52.
- Ritchie DF. 1999. Sprays for control of bacterial spot of peach cultivars having different levels of disease susceptibility. Fungic Nematic Tests. 54:63–64.
- Roselló M, Santiago R, Palacio-Bielsa A, García-Figueres F, Montón C, Cambra MA, López MM. 2012. Current status of bacterial spot of stone fruits and almond caused by Xanthomonas arboricola pv. pruni in Spain. J Plant Pathol. 94(1 Supplement):S1.15-S1.21.
- Schwarczinger I, Bozsó Z, Szatmári Á, Süle S, Szabó Z, Király L. 2017. First report of bacterial spot caused by Xanthomonas arboricola pv. prunion apricot in Hungary. Plant Dis. 101:1031.
- Shen YM, Huang TC, Chao CH, Liu HL. 2013. First report of bacterial spot caused by Xanthomonas arboricola pv. pruni on Japanese plum in Taiwan. Plant Dis. 97:835.
- Smith EF. 1903. Observations on a hitherto unreported bacterial disease, the cause of which enters the plant through ordinary stomata. Science. 17:456–457.
- Stefani E. 2010. Economic significance and control of bacterial spot/canker of stone fruits caused by Xanthomonas arboricola pv. pruni. J Plant Pathol. 92(1 Supplement):S1.99–S1.103.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tjou-Tam-Sin NNA, van de Bilt JLJ, Bergsma-Vlami M. 2012. First report of Xanthomonas arboricola pv. pruni in ornamental Prunus laurocerasus in the Netherlands. Plant Dis. 96:759.
- Yang L, Liu LJ, Li X. 2011. Status quo of production and exporting trades of peach and nectarines in China and its outlook for the near future. Agricult Outlook. 3:48–52. (in Chinese).