Abstract
White mould can affect the production of a wide range of economically important crops worldwide. The symptoms may be caused by several species, including Sclerotinia subarctica, a species mostly occurring in northern latitudes in sympatry with S. sclerotiorum. Although the two species are morphologically indistinguishable, S. subarctica was reported to have different climatic requirements for mycelial growth and production of apothecia. These differences may affect the precision of white mould risk prediction models that are based on the production of ascospores by S. sclerotiorum. To assess the presence of S. subarctica in France, we adapted a rapid PCR-based test to distinguish S. subarctica from other commonly found species of Sclerotinia. This test was used to characterize a collection of 969 Sclerotinia sp. isolates originating from various plants (bean, canola, carrot, lettuce, melon and witloof chicory), air and soil samples in different regions of France. One single isolate, collected from witloof chicory in northern France, was identified as S. subarctica. When genotyped with five microsatellite markers designed for S. sclerotiorum, this isolate had a haplotypic profile that was clearly distinct from the other isolates. The ITS sequence of this isolate was identical to those of isolates collected in northern Europe and Alaska. Koch’s postulates were verified. When inoculated on witloof chicory, the isolate identified as S. subarctica induced white mould symptoms. This study is the first to report the presence of S. subarctica south of the 51st parallel north and on witloof chicory.
Résumé
La pourriture blanche affecte la production d’un vaste panel de cultures économiquement importantes partout dans le monde. Les symptômes peuvent être causés par plusieurs espèces, dont S. subarctica, une espèce principalement présente sous les latitudes septentrionales en sympatrie avec S. sclerotiorum. Bien que les deux espèces ne puissent être distinguées sur la base de critères morphologiques, S. subarctica semble avoir besoin de conditions climatiques différentes pour sa croissance mycélienne et sa production d’apothécies. Ces différences peuvent affecter la précision des modèles de prédiction de risque de pourriture blanche qui sont basés sur la production d’ascospores par S. sclerotiorum. Pour évaluer la présence de S. subarctica en France, nous avons adapté un test PCR rapide pour distinguer S. subarctica des autres espèces de Sclerotinia communément rencontrées. Ce test a été utilisé pour caractériser une collection de 969 isolats de Sclerotinia sp. provenant de plantes (haricot, colza, carotte, laitue, melon, endive), d’air et de sol dans différentes régions de France. Un seul isolat, prélevé sur endive au nord de la France, a été identifié comme S. subarctica. Lorsque cet isolat a été genotypé avec 5 marqueurs microsatellites spécifiques de S. sclerotiorum, son profil haplotypique était clairement distinct de celui des autres isolats. La séquence ITS de cet isolat était identique à celle des isolats prélevés en Europe du Nord et en Alaska. Les postulats de Koch ont été vérifiés. L’isolat identifié comme S. subarctica a produit les symptômes de pourriture blanche lorsqu’il a été inoculé sur endive. Cette étude est la première à signaler la présence de S. subarctica au sud du 51e parallèle nord et sur endive.
Introduction7
White mould, caused by Sclerotinia sclerotiorum Whetz. and other species of Sclerotinia, can affect a wide range of cultivated and wild plant species and may cause annual economic losses estimated to reach millions of dollars worldwide (Boland & Hall, Citation1994; Purdy Citation1979; Petrofeza & Nasser, Citation2012). On diseased plants, the pathogen usually produces numerous sclerotia that have a potential of survival for several years in the soil. Through carpogenic germination, the sclerotia may produce apothecia which release large numbers of anemophilic ascospores. This airborne inoculum is believed to play a key epidemiological role in many crops, including canola, carrot, and lettuce (Abawi & Grogan, Citation1979; Bom & Boland, Citation2000; Clarkson et al., Citation2007; Parker et al., Citation2014). Biological control is increasingly used, but the management of white mould relies largely on chemical control (Derbyshire & Denton-Giles, Citation2016; Nicot et al., Citation2016), although fungicide applications are not always completely effective and the development of fungicide resistance is a common threat (Derbyshire & Denton-Giles, Citation2016). A way to optimize the use, and thus the efficiency, of disease control methods is to adjust interventions to the occurrence of epidemiological risk. Several risk prediction models have been developed and may be implemented in decision support systems for growers (Bom & Boland, Citation2000; Clarkson et al., Citation2007; Foster et al., Citation2011; Willbur et al., Citation2016). Such models generally predict the development of apothecia and the release of airborne ascospores on the basis of climatic parameters such as air or soil temperature, air or soil moisture levels and leaf wetness. Field implementation of such models may be complicated by the occurrence of strains of the pathogen with differing biological requirements (Clarkson et al., Citation2007). The situation could be further complicated by the fact that different species of Sclerotinia may be implicated in the development of white mould. This could be the case with a cryptic species living in sympatry with S. sclerotiorum, S. subarctica nom. prov. (previously named Sclerotinia sp. 1). First reported in Norway on wild dicotyledonous plants (Holst-Jensen et al., Citation1998), the latter species was found on cultivated crops in northern regions, including Alaska (Winton et al., Citation2006), Norway (Nordskog et al., Citation2014; Warmington, Citation2014; Brodal et al., Citation2016; Clarkson et al., Citation2016, Citation2017), Scotland (Clarkson et al., Citation2010, Citation2013, Citation2017; Warmington, Citation2014) and Sweden (Warmington, Citation2014; Clarkson et al., Citation2017). Compared with S. sclerotiorum, which can be found at all latitudes (Plantwise, Citation2017), S. subarctica appears to be confined to regions with cold weather. This geographic distribution is considered to reflect differences in biological requirements of the two species, including those pertaining to the low-temperature incubation period needed to trigger the carpogenic germination of sclerotia (Warmington Citation2014). Considering these biological differences, the accuracy of the risk-prediction models developed for S. sclerotiorum could be challenged if S. subarctica is abundantly present in the field.
The phenotypic differentiation of S. subarctica and S. sclerotiorum isolates is not easy. The two species are considered to produce similar white mould symptoms and to be morphologically indistinguishable (Holst-Jensen et al., Citation1998), although S. subarctica was found to produce fewer but slightly larger sclerotia in vitro than S. sclerotiorum in the UK (Clarkson et al., Citation2010). The two species can be clearly differentiated on the basis of the ITS sequence and that of the 5.8S rRNA gene (Holst-Jensen et al., Citation1998). Compared with S. sclerotiorum, S. subarctica was also found to lack an intron in the large subunit of rDNA, but the intron is also absent in S. minor isolates and thus this could not be used for the specific detection of S. subarctica. Several microsatellite markers have been reported to be present in S. subarctica but absent in S. sclerotiorum (Winton et al., Citation2007) and could offer a potential as a simple identification tool if their specificity can be further ascertained.
In France, white mould is very damaging on vegetable and arable crops and efforts are underway to provide risk prediction tools to help growers manage the disease. No information is available on the presence of S. subarctica on French crops, but its detection in nearby England prompted concerns about possible airborne dissemination across the Channel or from northern European countries. The present study was initiated to assess the possible occurrence of S. subarctica in France. To this end, a collection of isolates originating from different regions in France was examined, using molecular tools adapted from previously reported microsatellite markers.
Materials and methods
Fungal isolates
The study was carried out on a total of 969 isolates initially identified as S. sclerotiorum (based on morphological features) in the fungal collection of our laboratory. All isolates, except 11, were collected between 2011 and 2016. These isolates originated from diseased plants, air and field soil in France (). Among these, 311 isolates originated from sclerotia collected from diseased crops of bean (Phaseolus vulgaris L.), canola (Brassica napus L.), carrot (Daucus carota L.), endive (Cichorium endivia L.), flax (Linum usitatissimum L.), hemp (Cannabis sativa L.), lettuce (Lactuca sativa L.), melon (Cucumis melo L.), pea (Pisum sativum L.), tomato (Solanum lycopersicum L.) and witloof chicory (Cichorium intybus L.) and the weed Amaranthus sp. (). An additional 650 isolates originated from air and eight from soil ().
Table 1. Origin of the 969 Sclerotinia spp. isolates tested for possible identification as S. subarctica.
An isolate of S. subarctica collected from meadow buttercup in England, kindly provided by Dr J. Clarkson (Clarkson et al., Citation2013), was used as a reference. Isolates of S. trifoliorum and S. minor were kindly provided by GEVES (Beaucouzé, France) and by the IGEPP research unit of INRA (Le Rheu, France).
Specific molecular detection of S. subarctica
To devise a simple test to distinguish S. subarctica from S. sclerotiorum, two microsatellite loci were used among those reported by Winton et al. (Citation2007) to be specific for S. subarctica. These two loci, scsp1-ms01 and scsp1-ms08, were selected because of the non-overlapping size ranges reported for the amplicons (respectively 130–150 bp and 372–380 bp).
The specificity of these two microsatellite loci was verified for 13 Sclerotinia isolates (six of S. sclerotiorum, collected from bean, canola, carrot, lettuce, melon and witloof chicory; three of S. trifoliorum; three of S. minor; and one of S. subarctica). Six isolates of Botrytis cinerea were also included because this fungus is closely related to Sclerotinia sp. and has a comparable host range. All isolates were grown on PDA for 4 days at 21°C and the mycelium was collected from the plates and stored at −20°C before DNA extraction. Genomic DNA was extracted in 96-well plates from 100 mg aliquots of fresh frozen fungal material, following the Dneasy Plant extraction Kit protocol (Qiagen, ID:69181).
The PCR mix for the amplification of the microsatellite loci consisted of 2 uL of genomic DNA extract and 8 uL of a solution containing 1× QIAGEN Multiplex PCR Master Mix, 0.2 uM forward and reverse primers and 0.5× Q-solution (QIAGEN). PCRs were performed using the protocol for amplification of microsatellite loci (QIAGEN) with an initial denaturation step of 15 min at 95°C, followed by 30 cycles of 30 s at 94°C, 90 s at 55°C, and 60 s at 72°C, followed by a final step of 30 min at 60°C. The amplification products were visualized on an electrophoresis gel (1.5% agarose) amended with 1% ethidium bromide (30 min of migration at 80V). The bands corresponding to scsp1-ms01 and scsp1-ms08 were identified.
As the scsp1-ms08 pair of primers provided the strongest amplification, it was used to characterize the 963 additional field isolates of Sclerotinia sp., following the same PCR protocol as described above.
Verification of S. subarctica identity
Isolates putatively identified as S. subarctica with the test described above were further examined. Their ITS region was amplified, using ITS1 (5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ) and ITS4 (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) primers with the same protocol as described above, and the PCR products were sequenced (Genoscreen, Lille, France). The sequences were then compared to sequences of S. subarctica recorded in the GenBank data base.
A second verification of the taxonomic determination was carried out by genotyping all isolates in the collection with five additional microsatellite markers designed for S. sclerotiorum by Sirjusingh & Kohn (Citation2001). When tested on S. subarctica isolates, Winton et al. (Citation2006) showed that no amplification occurred for three of these loci (13–2, 55–4 and 110–4) and that the two others yielded alleles different from those expected for S. sclerotiorum (347 or 370 bp for locus 17–3 and 320 bp for locus 114–4). In the present study, markers were amplified with forward primers conjugated with the following fluorescent dyes: FAM for loci 13–2, 17–3 and 55–4, and HEX for loci 110–4 and 114–4 (MWG). Reverse primers did not carry any fluorescent dye. All markers were amplified at a hybridization temperature of 55°C, except 55–4 which was amplified at 60°C. To determine the size of the microsatellites, the PCR products were diluted and multiplexed prior to scanning with the help of an ABI 3730 sequencer (Applied Biosystems). In each well, 600LIZ was used as a standard size marker. GeneMapper software version 4.1 (Applied Biosystems) was then used for the microsatellite size analysis.
Koch’s postulates
Leaves of witloof chicory were inoculated with isolate SS_2428, following a method routinely used in our laboratory for pathogenicity tests with B. cinerea (Leyronas et al., Citation2015). In summary, the isolate was grown on PDA for 3 days at 21°C. Agar plugs (5 mm in diameter) were excised from the growing margin of the colony and placed in the centre of detached witloof chicory leaves (10 leaves). Ten control leaves received plugs of PDA without mycelium. The leaves were placed on humid absorbent paper in transparent polystyrene boxes incubated in a growth chamber at 21°C under a 10 h photoperiod (162 umol s−1 m−2) and examined daily for symptom development. This test was repeated twice independently.
Results
Sclerotinia subarctica identification with microsatellite markers
The primers for both microsatellite markers scsp1-ms-01 and scsp1-ms-08 amplified the DNA of the reference S. subarctica isolate, but not that of the six S. sclerotiorum, three S. minor, three S. trifoliorum and six B. cinerea isolates (). The scsp1-ms-08 pair of primers provided bands with a higher level of fluorescence and was then used to detect S. subarctica.
Fig. 1 Electrophoretic gel showing PCR products after amplification of microsatellite loci scsp1-ms-01 and scsp1-ms-08 (Winton et al., Citation2007), on Sclerotinia sp. and Botrytis cinerea isolates.
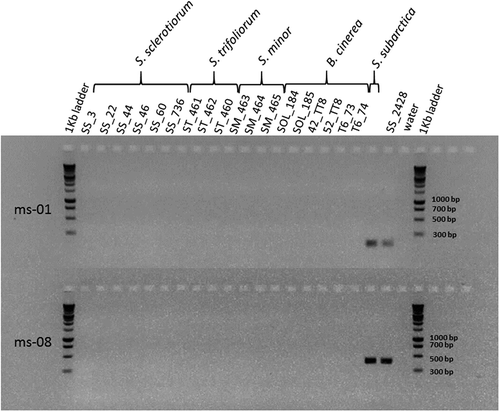
The entire collection of 969 field isolates of Sclerotinia spp. amplified the five microsatellites designed for S. sclerotiorum. All five markers were polymorphic with a number of alleles ranging from 6 to 19 (110–4 and 114–4 loci, respectively) (). One isolate (SS_2428) had a haplotypic profile that was clearly distinct from the others. It showed alleles at loci 114–4 and 17–3 that were not present in any other isolate: a 315 bp allele at locus 114–4 and a 369 bp allele at locus 17–3. Furthermore, there was no amplification for the three other loci (13–2, 55–4 and 110–4). The isolates of S. trifoliorum and S. minor also amplified two of the five microsatellites (loci 110–4 and 114–4).
Table 2. Polymorphism of five microsatellite markers among 968 isolates of Sclerotinia sp. and one isolate identified as S. subarctica.
With microsatellite ms-08 designed for S. subarctica, only one among all these isolates amplified a band in the size range of the scsp1-ms-08 locus. This isolate (SS_2428), collected in 2011 from witloof chicory stored in forcing trays in Arras (Lat. 50.28, Long. 2.78), also gave an amplification with the scsp1-ms-01 pair of primers (). For this isolate, the DNA amplification with ITS1 and ITS4 primers gave a 483 bp amplicon. BLASTn analysis showed that its sequence had 100% identity with that of isolates of S. subarctica collected from meadow buttercup in England deposited in GenBank (accession no. GU018183) (Clarkson et al., Citation2010), from turnip rape in Norway (no. KX929095.1) (Brodal et al., Citation2016), from vegetables in Alaska (formerly Sclerotinia sp. 1, no. EF091810.1, no. DQ329530.1, no. DQ329531.1 and no. DQ329532.1) (Winton et al., Citation2006) and from weeds in Norway (no. Z99677.1) (Holst-Jensen et al., Citation1998).
Induction of white mould symptoms on witloof chicory by S. subarctica
Four days after inoculation with mycelial plugs of SS_2428, all witloof chicory leaves showed brown lesions covered by white fluffy mycelium. The non-inoculated control leaves showed no symptom. After 6 days, black sclerotia were observed on inoculated leaves. These sclerotia were surface-sterilized in sodium hypochlorite, rinsed three times in sterile water and transferred to fresh PDA. They produced mycelium within 24 h and the resulting colonies developed white mycelium and produced new sclerotia after 6 days. These sclerotia were identical to those formed on witloof chicory and presented the same morphology and size range as those generally observed for S. sclerotiorum.
Discussion
This study showed that the amplification of microsatellite locus scsp1-ms-08 can provide a useful, rapid and economical tool to detect S. subarctica among isolates of Sclerotinia spp. This technique was cheaper than sequencing the ITS region and required half a day to identify S. subarctica isolates following DNA extraction compared with several days required for sequencing. The technique allowed the rapid screening of 969 Sclerotinia spp. isolates collected in France from different geographic regions and origins (air, soil) and from several host plants. As a result, the presence of S. subarctica is reported for the first time on witloof chicory, enlarging the known host range of this species. Until now, S. subarctica has been reported infecting wild and forage plants such as clover, dandelion (Taraxacum sp.), meadow buttercup (Ranunculus acris L.), northern wolfsbane (Aconitum septentrionale Koelle), yellow marsh marigold (Caltha palustris L.) (Holst-Jensen et al., Citation1998; Clarkson et al., Citation2010, Citation2013, Citation2017; Warmington, Citation2014) and on vegetables such as bean, cabbage, carrot, celery root, Jerusalem artichoke, lettuce, pea, potato, pumpkin, rutabaga and turnip rape (Winton et al., Citation2006; Nordskog et al., Citation2014; Warmington, Citation2014; Brodal et al., Citation2016; Clarkson et al., Citation2017). This host range is still quite modest compared with that of S. sclerotiorum (more than 400 species, Boland & Hall, Citation1994), but it is likely to continue increasing in future years, as suggested by infectivity tests carried out with S. subarctica isolates from meadow buttercup on additional plant species such as broccoli, canola and turnip (Clarkson et al., Citation2010; Taylor et al., Citation2015).
This study is also the first to report the presence of S. subarctica south of the 51st parallel north. Considering the observation of a single isolate (SS_2428) of this species among the 969 tested, S. subarctica is rare in French crops. The production of witloof chicory is concentrated in a region overlapping northern France (Boursies Lat. 50.13, Long. 3.01; Arras Lat. 50.28, Long. 2.78) and southern Belgium (Beitem Lat. 50.89, Long. 3.11; Herent Lat. 50.90, Long. 4.67), subjected to a temperate oceanic climate (Peel et al., Citation2007). This region is located ~500 km from Michaelchurch Escley, Herefordshire, England, the closest area to where S. subarctica was reported (Clarkson et al., Citation2013, Citation2017), and ~700 km from areas in Scotland and 1000 km from Scandinavia where the fungus is abundant (Warmington, Citation2014; Brodal et al., Citation2016; Clarkson et al., Citation2017). As fungal spores are known to survive high altitudes in the atmosphere (Meier et al., Citation1933) and long distance dispersal (Brown & Hovmøller, Citation2002), occasional introductions of airborne inoculum of S. subarctica from those regions should not be excluded. Other means of dissemination related to human activities, such as movement of plants and seeds, should also be considered. Aside from SS_2428, all other isolates examined in the present study from air samples of witloof chicory fields (114 isolates collected between 2014 and 2016) and from plants (27 collected between 2012 and 2015) were identified as S. sclerotiorum on the basis of their morphological traits and the amplification of the five microsatellite markers (Sirjusingh & Kohn, Citation2001). Further sampling and characterization of Sclerotinia spp. isolates in this region over the ensuing years would help to determine if S. subarctica is established there, possibly through an acclimation to warmer climate, or if its present detection is the result of an accidental and rare introduction.
Acknowledgements
The authors greatly thank Dr John Clarkson (University of Warwick, UK) for providing a reference isolate of S. subarctica. Isolates from canola were kindly provided by Terres Inovia (Grignon, France) and those of S. trifoliorum and S. minor were kindly provided by GEVES (Beaucouzé, France) and INRA research unit IGEPP (Le Rheu, France). We thank all the field experimenters of the technical institutes (CTIFL, ACPEL, APEF, CEFEL, INVENIO, SILEBAN, Terres Inovia UNILET) who collaborated in the project and who collected the isolates used in the present study.
Additional information
Funding
References
- Abawi GS, Grogan RG. 1979. Epidemiology of diseases caused by Sclerotinia species. Phytopathology. 69:899–904.
- Boland GJ, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol. 16:93–108.
- Bom M, Boland GJ. 2000. Evaluation of disease forecasting variables for sclerotinia stem rot (Sclerotinia sclerotiorum) of canola. Can J Plant Sci. 80:889–898.
- Brodal G, Warmigton R, Grieu C, Ficke A, Clarkson JP. 2016. First report of Sclerotinia subarctica nom. prov. (Sclerotinia sp. 1) causing stem rot on turnip rape (Brassica rapa subsp. oleifera) in Norway. Plant Dis. 101:386–396.
- Brown JKM, Hovmøller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 297:537–541.
- Clarkson JP, Carter HE, Coventry E. 2010. First report of Sclerotinia subarctica nom. prov. (Sclerotinia species 1) in the UK on Ranunculus acris. Plant Pathol. 59:1173–1273.
- Clarkson JP, Coventry E, Kitchen J, Carter HE, Whipps JM. 2013. Population structure of Sclerotinia sclerotiorum in crop and wild hosts in the UK. Plant Pathol. 62:309–324.
- Clarkson JP, Phelps K, Whipps JM, Young CS, Smith JA, Watling M. 2007. Forecasting sclerotinia disease on lettuce, a predictive model for carpogenic germination of Sclerotinia sclerotiorum sclerotia. Phytopathology. 95:621–631.
- Clarkson JP, Warmington R, Nordskog B. 2016. Population structure of Sclerotinia subarctica in England, Scotland and Norway. Phytopathology. 106(Suppl):139–149.
- Clarkson JP, Warmington R, Walley PG, Denton-Giles M, Barbetti MJ, Brodal G, Nordskog B. 2017. Population structure of Sclerotinia subarctica and Sclerotinia sclerotiorum in England, Scotland and Norway. Front Microbiol. 8. doi:10.3389/fmicb.2017.00490
- Derbyshire MC, Denton-Giles M. 2016. The control of sclerotinia stem rot on oilseed rape (Brassica napus), current practices and future opportunities. Plant Pathol. 65:859–877.
- Foster AJ, Kora C, McDonald MR, Boland G. 2011. Development and validation of a disease forecast model for sclerotinia rot of carrot. Can J Plant Pathol. 33:187–201.
- Holst-Jensen A, Vaage M, Schumacher T. 1998. An approximation to the phylogeny of Sclerotinia and related genera. Nord J Bot. 18:705–719.
- Leyronas C, Bryone F, Duffaud M, Nicot PC. 2015. Assessing host specialization of Botrytis cinerea on lettuce and tomato by genotypic and phenotypic characterization. Plant Pathol. 64:119–127.
- Meier FC, Stevenson JA, Charles VK. 1933. Spores in the upper air. Phytopathology. 23:23–24.
- Nicot PC, Avril F, Duffaud M, Leyronas C, Troulet C, Villeneuve F, Bardin M. 2016. Are there regional differences in the susceptibility of Sclerotinia sclerotiorum strains to Coniothyrium minitans? IOBC-WPRS Bull. 117:83–87.
- Nordskog B, Eikemo H, Gauslaa E, Le VH, Warmington R, Clarkson JP. 2014. Distribution and prevalence of Sclerotinia sclerotiorum and S. subarctica in Norwegian lettuce. Phytopathology. 104(Suppl):86–96.
- Parker ML, McDonald MR, Boland G. 2014. Assessment of spatial distribution of ascospores of Sclerotinia sclerotiorum for regional disease forecasting in carrots. Can J Plant Pathol. 36:438–446.
- Peel MC, Finlayson BL, McMahon TA. 2007. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci. 11:1633–1644.
- Petrofeza S, Nasser LCB. 2012. Case study: Sclerotinia sclerotiorum: Genetic diversity and disease control. In: Caliskan M, editor. The molecular basis of plant genetic diversity. Rijeka: InTech; p. 276–296. doi:10.5772/33780
- Plantwise. 2017. Sclerotinia sclerotiorum disease distribution map. [accessed 2017 Mar 8]. http://www.plantwise.org/knowledgebank/pwmap.aspx?speciesid=39097&loc=global.
- Purdy LH. 1979. Sclerotinia sclerotiorum - history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology. 69:875–880.
- Sirjusingh C, Kohn LM. 2001. Characterization of microsatellites in the fungal plant pathogen, Sclerotinia sclerotiorum. Mol Ecol Notes. 1:267–269.
- Taylor A, Coventry E, Jones JE, Clarkson JP. 2015. Resistance to a highly aggressive isolate of Sclerotinia sclerotiorum in a Brassica napus diversity set. Plant Pathol. 64:932–940.
- Warmington RJ 2014. Pathogen diversity, epidemiology and control of Sclerotinia disease in vegetable crops [dissertation]. UK: University of Warwick.
- Willbur J, Lucas H, Mueller B, Chapman S, Kabbage M, Smith D. 2016. Validation and refinement of a predictive model for Sclerotinia sclerotiorum apothecial development in soybean fields. Phytopathology. 106(Suppl):5–6.
- Winton LM, Krohn AL, Leiner RH. 2007. Microsatellite markers for Sclerotinia subarctica nom. prov., a new vegetable pathogen of the High North. Mol Ecol Notes. 7:1077–1079.
- Winton LM, Leiner RH, Krohn AL. 2006. Genetic diversity of Sclerotinia species from Alaskan vegetable crops. Can J Plant Pathol. 28:426–434.
