Abstract
Potato leaves with symptoms of foliar diseases were collected from eight different geographical areas in southern China. A total of 109 Alternaria isolates were recovered and identified to three species according to morphological characteristics, nucleotide sequencing and PCR-RFLP analysis. Three Alternaria species were recovered, of which A. tenuissima was the most common, followed by A. alternata and A. solani. Pathogenicity tests indicated that all species caused foliar symptoms on potato cultivar ‘Favorite’. Compared with earlier results reported in 2010–2013 in northern China, the proportion of A. alternata isolates was notably increased, while the number of A. solani isolates was lower. This study provides a survey of the Alternaria species composition associated with potato foliar diseases in southern China.
Résumé
Des feuilles de pomme de terre affichant des signes de maladies foliaires ont été collectées dans huit régions différentes du sud de la Chine. En tout, 109 isolats d’Alternaria ont été recueillis et identifiés à 3 espèces en fonction de leurs caractéristiques morphologiques, du séquençage des nucléotides et de l’analyse PCR-RFLP. Trois espèces d’Alternaria ont été récupérées, parmi lesquelles A. tenuissima était la plus courante, suivie d’A. alternata et d’A. solani. Des tests de pathogénicité ont indiqué que toutes les espèces causaient des symptômes de maladies foliaires sur le cultivar de pomme de terre ‘Favorite’. Comparativement aux résultats précédents provenant du nord de la Chine en 2010–2013, la proportion d’isolats d’A. alternata s’était substantiellement accrue, tandis que le nombre d’isolats d’A. solani avait diminué. Cette étude passe en revue la composition des espèces d’Alternaria associées aux maladies foliaires de la pomme de terre dans le sud de la Chine.
Introduction
Potato (Solanum tuberosum L.) is one of the most important food crops in the world (Park et al., Citation2009) and is affected by many foliar diseases, including early blight, leaf spot and brown spot in China and elsewhere (Ardestani et al., Citation2010). Ten species of Alternaria have been implicated to cause foliar diseases of potato worldwide. These include A. solani (Leiminger et al., Citation2015), A. alternata, A. tenuissima, A. dumosa, A. arborescens, A. infectoria (Ardestani et al., Citation2010), A. grandis (Rodrigues et al., Citation2010), A. interrupta (Taheri et al., Citation2009), A. longipes (Shoaib et al., Citation2014) and A. arbusti (Tymon & Johnson, Citation2014; Tymon et al., Citation2016). From 2010 to 2013, a total of 511 Alternaria isolates comprising three species were obtained from diseased potato leaves sampled from regions of northern China, and A. tenuissima was the most prevalent (Zheng et al., Citation2015). Knowledge of which Alternaria species are associated with potato foliar diseases nationwide may be important in designing both chemical and biological management schemes in China. Thus, the aim of this study was to identify the Alternaria species composition causing potato foliar diseases in southern China.
Materials and methods
Isolation and characterization of Alternaria isolates
In May of 2014 and 2015, diseased potato leaves with symptoms of early blight, leaf spot and brown spot were sourced from eight provinces, autonomous regions or municipalities in southern China. Diseased leaf tissues were cut into 3–5 mm pieces and surface-sterilized in 0.3% sodium hypochlorite for 2 min, rinsed in sterile distilled water three times and placed onto Petri dishes containing potato dextrose agar (PDA) amended with streptomycin sulphate (50 mg·L−1). A total of 109 pure cultures purified by single spore method and confirmed microscopically to be Alternaria species were obtained and transferred onto PDA slants. Colony characters were determined after these isolates were incubated on PDA and potato carrot agar (PCA) plates (Zheng et al., Citation2015) at 25°C for 7 days. PDA plates were kept in darkness while PCA plates were kept 40 cm below cool white fluorescent bulbs with 8 h/16 h periods of light/dark (Andersen et al., Citation2001; Simmons, Citation2007).
Pathogenicity test
To determine the pathogenicity of the 70 representative Alternaria isolates, potato (cv. ‘Favorite’) was grown in a greenhouse for 45 days and maintained at 22–25°C with 85–90% RH. For preparation of spore suspensions, A. tenuissima and A. alternata were incubated on PDA plates at 25°C for 7 days; while A. solani was incubated on V8 juice agar plates at 22°C for 7 days in the dark and then transferred to a growth chamber with a 16 h light cycle at 20°C for 2 days (Langsdorf et al., Citation1990). For each Alternaria isolate tested, 10 detached apical leaflets from 45-day-old potato plants, replicated three times, were inoculated with a conidial suspension of 106 conidia·mL−1. Leaflets inoculated with sterile distilled water served as a control. All inoculated leaflets were placed on Petri dishes lined with moist filter paper in plastic boxes and incubated in a growth chamber at 25°C and 90% RH with a 12 h photoperiod. After incubation for 7 days, pathogenicity of the Alternaria isolates was assessed by presence or absence of symptoms occurring on the leaflets. Disease severity (DS) was scored on a 4-point rating system (Pryor & Michailides, Citation2002), and disease index (DI) was calculated to compare the pathogenicity of the three Alternaria species (Zheng et al., Citation2015). Fungi were re-isolated from the infected leaflets and confirmed by morphological characteristics and molecular methods to fulfil Koch’s postulates. The experiment was repeated twice for confirmation of the results.
DNA sequencing and phylogenetic analysis
In order to confirm the results from morphological studies, 70 Alternaria isolates representing different geographical origins or Alternaria species were grown on PDA plates and genomic DNA was extracted using the method described by Lee & Taylor (Citation1990). Partial coding sequences of the histone 3 gene were amplified using primer sets H3-1a (5′-ACT AAG CAG ACC GCC CGC AGG-3′) and H3-1b (5′-GCG GGC GAG CTG GAT GTC CTT-3′) (Glass & Donaldson, Citation1995) for molecular identification of the isolates according to previously reported procedures (Zheng et al., Citation2015). The PCR products were purified and sequenced by Beijing Sunbiotech Co. Ltd. Blast search at the National Centre for Biotechnology Information (NCBI) was performed for each nucleotide sequence. Sequence alignment was done using Clustal W program (version 1.83) (Thompson et al., Citation1994) with manual adjustment. Phylogenetic analysis of the Alternaria isolates was carried out by MEGA 5 program version 5.2.2 (http://www.megasoftware.net/) using the neighbour-joining (NJ) method. Alternaria infectoria was used as the out-group taxon (Peever et al., Citation2004).
PCR-RFLP analysis of histone 3 gene
For PCR-RFLP analysis, two restriction enzymes (TaqI and HapII; TaKaRa, Japan) were chosen to digest the partial coding sequence of histone 3 gene of the three Alternaria species. Restriction analyses of the amplification products were performed as described by Pryor & Michailides (Citation2002). Digestion products were resolved in 2% agarose gel in TBE buffer and visualized by UV illumination after staining in ethidium bromide. The size of DNA fragments was determined in comparison with a 100 bp ladder. PCR-RFLP analysis was performed twice to confirm reproducibility of amplification and digestion products.
Results and discussion
Characterization of the Alternaria isolates
Based on morphological characters, a total of 109 Alternaria isolates were obtained from potato leaves with foliar symptoms in southern China, which were grouped into three morphological groups according to the original description of Alternaria species (Simmons, Citation2007). Amongst these isolates, A. tenuissima was the most prevalent (58 isolates, 53.2%), followed by A. alternata (48 isolates, 44.0%), and A. solani (three isolates, 2.8%).
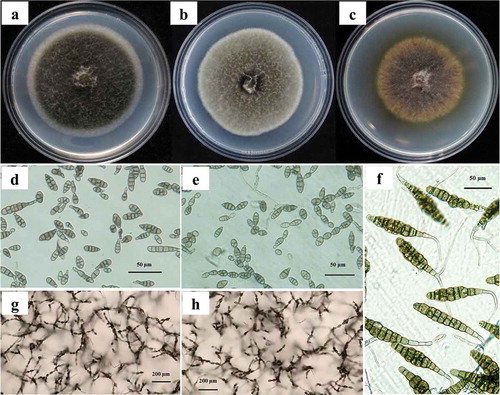
Colonies of isolates in the A. tenuissima group were cottony, greyish green to olive brown, often with a very thin white margin (). The conidial chains were up to 12 conidia in length with uncommon secondary chains (). Conidia were 26.5–45.3 × 11.2–16.0 µm and typically ovoid to obclavate in shape with 1 to 6 transverse septae and 0 to 2 longitudinal septae (). Colonies of isolates in the A. alternata group were dense and dark grey, usually with a prominent white margin (). The conidial chains were 8 to 10 conidia in length with numerous secondary and occasionally tertiary chains (). Conidia were ovoid, obpyriform to ellipsoid, 26.0–39.5 × 6.7–11.0 μm in size, with 3–7 transversal and 0–3 longitudinal septae (). Colonies of isolates in the A. solani group were dense dark grey to black with sparse aerial mycelia (), and produced simple conidiophores bearing dark conidia with 9–11 transverse septae and 1–2 longitudinal septae. Conidia with one beak were long-ovoid or long-ellipsoid, 107.5–115.0 × 18.0–25.0 μm in size with a beak 80.5–110.0 μm in length. Conidia with 2–3 beaks were shorter, 82.3–104.0 × 16.5–20.3 μm with an initial beak 64.8–85.5 μm long and a second beak 58.5–86.3 μm long (). The morphological characters of the three Alternaria species were the same as those reported in the literature (Simmons, Citation2007).
Fig. 1 (Colour online) Colonies, conidia, and sporulation patterns of three Alternaria species after 7 days of growth on PDA or PCA plates. a–c, Colonies of isolates representing A. tenuissima, A. alternata, A. solani on PDA plates. d–f, Conidia of isolates representing A. tenuissima, A. alternata, A. solani on PCA plates, scale bars: 50 μm. g–h, sporulation patterns of isolates representing A. tenuissima, A. alternata on PCA plates, scale bars: 200 μm.
Pathogenicity test
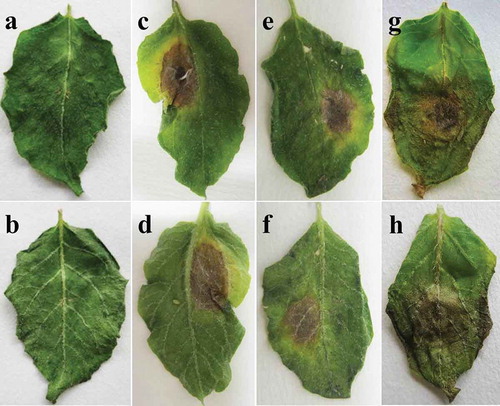
Inoculated potato leaflets developed disease symptoms such as round or oval, light to dark brown lesions when inoculated with spore suspensions of the 70 Alternaria isolates. In comparing lesion development among the different Alternaria species, no significant differences in disease incidence could be detected; however, the disease index of potato leaves inoculated with A. solani was significantly higher than the other two species (P < 0.05). None of the control leaves inoculated with sterile distilled water developed disease symptoms (). The pathogen was re-isolated and found to be identical to the original isolate. Results revealed that the three Alternaria species were the causal agents of potato foliar disease.
Fig. 2 (Colour online) Pathogenicity of the representative isolates of three Alternaria species on detached potato leaflets. a-b, sterile water controls. c–d, A. tenuissima. e–f, A. alternata. g–h, A. solani. The experiment was conducted using detached apical leaflets from 45-day-old plants of potato cv. Favorite. A spore suspension of 106 conidia·ml−1 was inoculated on the upper surface of each leaflet (one point per leaflet).
DNA sequencing and phylogenetic analysis
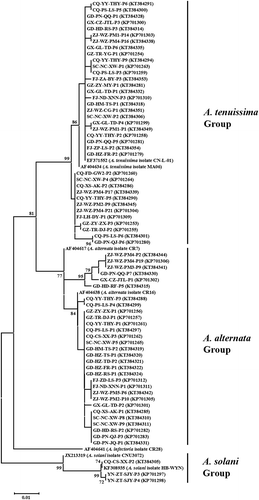
PCR amplification of the histone 3 gene generated predicted sizes of 546 bp, 440 bp and 489 bp fragments for isolates in the morphological A. tenuissima group (A-ten), A. alternata group (A-alt) and A. solani group (A-sol), respectively. BLAST searches revealed that the histone 3 genes of the Alternaria isolates in Group A-ten, A-alt and A-sol shared 99–100% similarity with those of A. tenuissima, A. alternata and A. solani in the GenBank database. The sequences of the tested Alternaria isolates were deposited in GenBank (Supplementary Table 1). All these Alternaria isolates were grouped into three distinct phylogenetic clades representing A. tenuissima, A. alternata and A. solani species on the basis of their sequences of histone 3 gene ().
Fig. 3 Phylogenetic tree constructed based on the partial coding sequences of histone 3 gene of 37 A. tenuissima isolates, 30 A. alternata isolates, three A. solani isolates, as well as seven reference sequences retrieved from GenBank. The tree was constructed by neighbour joining using the Kimura two-parameter distance method. Bootstrap values (in percentage) above 70 from 1000 pseudo-replicates are shown for major lineages within the tree.
PCR-RFLP analysis of histone 3 gene
The bands generated by restriction digestion were of the predicted sizes obtained with DNAMAN restriction analysis. Direct digestion of the PCR products of the histone 3 genes with enzyme TaqI generated reproducible species-specific restriction fragments of 331 and 118 bp for A. tenuissima, 225 and 118 bp for A. alternata and 274 and 118 bp for A. solani. The short fragments of 73 and 24 bp, which were theoretically generated from the three species with TaqI digestion, were not visualized due to the limited resolution of electrophoresis in 2.0% agarose gel. Direct digestion with enzyme HapII cleaved the PCR products into 256 and 198 bp fragments for A. tenuissima isolates, 256 and 151 bp fragments for A. alternata isolates, 256 and 141 bp fragments for A. solani isolates. PCR-RFLP analysis of the histone 3 gene successfully differentiated the three Alternaria species, especially A. tenuissima and A. alternata, which could not be resolved by the PCR-RELP of IGS region (Pryor & Michailides, Citation2002).
Alternaria species are usually plant pathogenic and may damage plants such as potato, sunflower, melon, watermelon in the field according to former surveys (Wang et al., Citation2014; Zhao et al., Citation2016a, Citation2016b). An accurate understanding of the pathogen population in different regions of China is crucial for suitable fungicide selection and efficient disease control. From 2010 to 2013, a total of 511 Alternaria isolates consisting of A. tenuissima, A. alternata and A. solani were obtained from diseased potato leaves sampled in geographical origins of northern China (Zheng et al., Citation2015). In this study, A. tenuissima was also confirmed as the dominant species causing potato foliar diseases in southern China. However, the proportion of A. alternata isolates was increased, which was closer to that of A. tenuissima isolates, and the number of A. solani isolates was reduced. Proportional changes among the Alternaria species associated with potato foliar diseases might be caused by differences in temperature and humidity, growing conditions, as well as the diversity of crop varieties among the different regions. According to the present results, both of the most common species should be treated as the major target pathogens in decision on management strategies in southern China.
Supplemental Table 1
Download MS Word (95 KB)Acknowledgements
This work was supported by Chinese Universities Scientific Fund (2015NX005). Mention of trade names or commercial products in this report is solely for the purpose of providing specific information and does not imply recommendation or endorsement.
Supplemental material
Supplemental data for this article can be accessed https://doi.org/10.1080/07060661.2018.1459850.
Additional information
Funding
References
- Andersen B, Krøger E, Roberts RG. 2001. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol Res. 105:291–299.
- Ardestani ST, Sharifnabi B, Zare R, Moghadam AA. 2010. New Alternaria species associated with potato leaf spot in various potato growing regions of Iran. Iranian J Plant Pathol. 45:83–86.
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb. 61:1323–1330.
- Langsdorf G, Furuichi N, Doke N, Nishimura S. 1990. Investigations on Alternaria solani infections: detection of alternaric acid and a susceptibility-inducing factor in the spore-germination fluid of A. solani. J. Phytopathol. 128:271–282.
- Lee SB, Taylor JW. 1990. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. San Diego (CA): Academic Press; p. 282–287.
- Leiminger J, Bassler E, Knappe C. 2015. Quantification of disease progression of Alternaria spp. on potato using real-time PCR. Eur J Plant Pathol. 141:295–309.
- Park TH, Vleeshouwers VGAA, Jacobsen E, Van Der Vossen E, Visser RGF. 2009. Molecular breeding for resistance to Phytophthora infestans (Mont.) de Bary in potato (Solanum tuberosum L.): a perspective of cisgenesis. Plant Breed. 128:109–117.
- Peever TL, Su G, Carpenter-Boggs L, Timmer LW. 2004. Molecular systematics of citrus-associated Alternaria species. Mycologia. 96:119–134.
- Pryor BM, Michailides TJ. 2002. Morphological, pathogenic, and molecular characterization of Alternaria isolates associated with Alternaria late blight of pistachio. Phytopathology. 92:406–416.
- Rodrigues TTMS, Berbee ML, Simmons EG, Cardoso CR, Reis A, Maffia LA, Mizubuti ESG. 2010. First report of Alternaria tomatophila and A. grandis causing early blight on tomato and potato in Brazil. New Dis Rep. 22:28.
- Shoaib A, Akhtar N, Akhtar S, Hafeez R. 2014. First report of Alternaria longipes causing leaf spot of potato cultivar Sante in Pakistan. Plant Dis. 98:1742.
- Simmons EG. 2007. Alternaria: an identification manual. CBS biodiversity series 6. Utrecht (the Netherlands): CBS Fungal Biodiversity Centre.
- Taheri AS, Sharif NB, Zare R, Abbasi MA. 2009. Alternaria interrupta, a new pathogen causing potato early blight in Iran. Rostaniha. 10:72–73.
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Tymon LS, Cummings TF, Johnson DA. 2016. Pathogenicity and aggressiveness of three Alternaria spp. on potato foliage in the U.S. Northwest. Plant Dis. 100:797–801.
- Tymon LS, Johnson DA. 2014. Fungicide resistance of two species of Alternaria from potato in the Columbia Basin of Washington. Plant Dis. 98:1648–1653.
- Wang TY, Zhao J, Sun P, Wu XH. 2014. Characterization of Alternaria species associated with leaf blight of sunflower in China. Eur J Plant Pathol. 140:301–315.
- Zhao J, Bao SW, Ma GP, Wu XH. 2016a. Characterization of Alternaria species associated with muskmelon foliar diseases in Beijing municipality of China. J Gen Plant Pathol. 82:29–32.
- Zhao J, Bao SW, Ma GP, Wu XH. 2016b. Characterization of Alternaria species associated with watermelon leaf blight in Beijing municipality of China. J Plant Pathol. 98:135–138.
- Zheng HH, Zhao J, Wang TY, Wu XH. 2015. Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathol. 64:425–433.
