Abstract
Severe blight and necrotic symptoms were observed on a 3-year-old alfalfa crop cultivar ‘Blue Jay’ at Indian Head, Saskatchewan during the summer of 2017. Symptoms appeared on blossoms, leaves and stems, and most of the infected plants were severely defoliated. The objective of this study was to isolate and identify the causal agent of the disease. Isolations from diseased alfalfa plants showing visible blight and necrotic symptoms collected from the infected field were made on potato dextrose agar (PDA). Morphological observations of the developing fungal colonies on PDA showed the presence of dark grey to dark green mycelium and conidia borne in chains with vertical and transverse septa, indicating the presence of the fungus Alternaria alternata. Molecular analysis of the ITS-5.8S region of rDNA (541 bp) and the TEF-1α gene region (606 bp) of the fungal isolates from alfalfa confirmed their identity as A. alternata. The pathogenicity of representative isolates was confirmed on alfalfa ‘Blue Jay’ and ‘Spredor 4’ plants in growth chamber tests. Necrotic and blight symptoms like those observed in the field appeared on leaves and stems followed by defoliation. To our knowledge, this is the first report of A. alternata causing a foliar disease on alfalfa in Canada.
Résumé
À l’été de 2017, de graves symptômes associés au mildiou et à la nécrose ont été observés sur des cultures de trois ans de luzerne du cultivar ‘Blue Jay’ à Indian Head, en Saskatchewan. Les symptômes sont apparus sur les fleurs, les feuilles et les tiges, et la plupart des plants infectés étaient gravement défoliés. Le but de cette étude était d’isoler et d’identifier l’agent causal de la maladie. Les isolements des plants de luzerne affichant des symptômes visibles de mildiou et de nécrose, collectés dans le champ infecté, ont été faits sur de la gélose dextrosée à la pomme de terre. Les observations morphologiques du développement des colonies fongiques sur la gélose ont révélé des chaînes de mycélium et de conidies gris foncé à vert foncé possédant des cloisons verticales et transversales, indiquant qu’il s’agissait du champignon Alternaria alternata. L’analyse moléculaire de la région de l’ITS-5,8S de l’ADNr (541 bp) et de la région du gène codant pour le TEF-1α (606 bp) des isolats fongiques de la luzerne a confirmé leur identité en tant qu’A. alternata. La pathogénicité d’isolats représentatifs a été confirmée pour les cultivars ‘Blue Jay’ et 'Spredor 4' cultivés au cours de tests effectués en chambre de croissance. Des symptômes de nécrose et de mildiou, comme ceux observés au champ, sont d’abord apparus sur les feuilles et les tiges, puis il y a eu défoliation. À notre connaissance, il s’agit du premier rapport d’une maladie foliaire de la luzerne causée par A. alternata au Canada.
Introduction
Alfalfa (Medicago sativa L.) is an important crop widely grown for forage and hay throughout the world (Stuteville & Erwin, Citation1990). This deep-rooted perennial legume can play an important role in soil conservation and can increase soil nitrogen. Alfalfa is prone to attack by a number of foliar diseases that could limit its production by reducing yield and forage quality (Stuteville & Erwin, Citation1990). Most of the foliar diseases affecting alfalfa are known to be caused by fungal pathogens (Stuteville & Erwin, Citation1990; Nutter et al., Citation2002). Spring black stem and leaf spot, summer black stem and leaf spot, common leaf spot, Leptosphaerulina leaf spot, and Stemphylium leaf spot are common fungal diseases of alfalfa. Stemphylium leaf spot occurs in most areas where alfalfa is grown, with Stemphylium botryosum as the predominant pathogen (Cowling et al., Citation1981) and S. globuliferum as a minor pathogen (Samac et al., Citation2014).
Leaf spot and blight symptoms (Fig. 1a) were observed on a 3-year-old alfalfa ‘Blue Jay’ field in Indian Head, Saskatchewan during the summer of 2017. These plots had been established in June 2014 and are part of the Agriculture and Agri-Food Canada seed-increase programme and serve as a seed source to growers. It is worth noting that there were no other alfalfa fields adjacent to the affected field within a 500-m radius. The affected alfalfa field was previously under oats or green fallow since 2010. The plots were sprayed with the insecticide Matador (Lambda-cyhalothrin) in late June 2017. Disease symptoms appeared in early July and spread very rapidly, covering the whole field. Blight and necrotic symptoms were visible on leaves () and stems (). Blossoms were also infected and covered with brownish and blackish sooty material. Most of the infected plants were severely defoliated, and the affected field showed more than 80–90% disease incidence. The objective of this study was to isolate, characterize and identify the causative agent associated with the blight and necrotic symptoms on the alfalfa plants, and confirm its pathogenicity.
Fig. 1 (Colour online) Symptoms of Alternaria blight on alfalfa and morphological features of Alternaria alternata. a, Affected alfalfa ‘Blue Jay’ field; b, Necrotic and blight symptoms on foliage; c, Necrotic lesions on leaf; d, Colony of A. alternata after 7 days of growth at 25°C; e, Colony after 21 days of growth on PDA; f,,Mycelia and conidia of A. alternata; g, Chains of conidia of A. alternata; h, Necrotic symptoms on ‘Spredor 4’ and ‘Blue Jay’ plants inoculated with A. alternata; i, Necrotic lesion on inoculated alfalfa stem.

Materials and methods
Sampling and pathogen isolation
Three symptomatic plants were randomly collected from a 3-year-old alfalfa ‘Blue Jay’ field with necrotic and blight symptoms in 2017 and shipped to the Plant Pathology laboratory at Kentville Research and Development Centre. The infected tissues were examined under a compound microscope to observe the presence of pathogen propagules or fruiting structures. Isolations were made from blossom, leaf and stem tissues by cutting 1-cm long sections of tissue with a sterile scalpel, surface-sterilizing by immersion in a 30% (50% for stem tissues) (v/v) bleach solution for 1.5–2 min, followed by two rinses in sterile-distilled water (SDW). The tissue pieces were then placed onto potato dextrose agar (PDA, Difco, USA) plus chloramphenicol (80 mg L−1). Petri dishes were incubated for 7 days at 22 ± 2°C on a laboratory bench and examined daily to check for growing colonies. A similar type of fungal colony consistently developed from all three tissue sources and they were transferred onto new PDA dishes. Fungal isolates were maintained as stock cultures on PDA slants at 4°C.
Morphological characterization
A total of six (two/tissue) fungal isolates were selected for further comparisons of growth and morphological features. All isolates were grown on PDA for 7 days at 25°C and colony morphology (colour, shape) and characteristics of conidiophores (colour, size) and conidia were examined. The colour, shape and size of at least 30 conidia for each of the fungal isolates were recorded. Based on the similar morphological results obtained for all fungal isolates, three representative isolates – one each from blossom (17–03), leaf (17–06) and stem (17–02) tissues – were selected for pathogenicity tests on alfalfa plants and molecular identification. All three isolates were further purified by single conidium transfer on fresh PDA.
Pathogenicity tests
Conidial production was induced as described by Shahin & Shepard (Citation1979) with some modifications. In brief, the actively growing mycelium from the edges of 5-day-old PDA cultures were cut in 1-cm squares, any fluffy aerial mycelia was removed with a sterile scalpel, and the squares were placed on water agar (WA) plus calcium carbonate and 2 mL sterile distilled water was placed on top of the plugs. Petri dishes were incubated in the dark at 20°C and spores from square plugs and WA were harvested after 24-48 h by filtering through four layers of sterile cheesecloth. Pathogenicity of three isolates was tested on alfalfa ‘Blue Jay’ and ‘Spredor 4’ plants grown in a greenhouse potting mix in 12-cm diameter pots for 2 and 4 months, respectively. The greenhouse potting mix was prepared by mixing 85 L peat moss, 10 kg perlite, 20 L sand, 1 kg dolomitic limestone and 1 kg starter fertilizer [2-3-6] in a cement mixer. There were two replicate pots for each fungal isolate, containing 12–15 plants. Plants were inoculated by spraying both upper and lower sides of leaves with a freshly prepared spore suspension of each isolate at 104–105 conidia mL−1 with an atomizer (Devilbiss Healthcare, Somerset, PA). Inoculated plants were initially kept under mist conditions at >90% humidity in a growth chamber set at 22°C, and then maintained at 22°C/20°C (day/night) with 12 h photoperiod and continuous mist conditions. Plants were watered as required. The experiment was repeated twice. The symptoms on inoculated plants were assessed over a period of 3 weeks. To fulfil Koch’s postulates, inoculated leaf tissues with necrotic and blight-like symptoms were used to re-isolate the pathogen on PDA, as described above for fungal isolation. The morphological characteristics of the re-isolated cultures were then compared with those of the original culture. Leaf tissues from non-inoculated control alfalfa plants were also plated on PDA to ensure that the healthy plants used for the pathogenicity tests were not infected.
Molecular characterization and phylogenetic analysis
All three isolates were further characterized by PCR amplification of the internal transcribed spacer (ITS) region of 5.8 S ribosomal DNA (rDNA) and the translation elongation factor 1 alpha (TEF-1α) region. All isolates were grown on PDA for 7 days at 22 ± 2°C and mycelia and conidia were harvested by scraping the agar surface with a sterile scalpel. Genomic DNA was isolated using an E.Z.N.A. SP Fungal DNA Mini Kit (Omega Bio-Tek) following the manufacturer’s instructions. The ITS region of rDNA was amplified using universal primer pairs ITS1 (5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ) and ITS4 (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) (White et al., Citation1990). The TEF-1α region was amplified using primer pairs EF1-up (5ʹ-TCGTYATCGGCCACGTC-3ʹ) and EF1-low (5ʹ-GARGTACCAGTSATCATGTTCTT-3ʹ) (Al-Mughrabi et al., Citation2013). PCR amplification was performed in a 50 µL reaction mixture using 4 µL (5 ng µL−1) template DNA, 5 µL 10× PCR buffer (750 mM Tris-HCl (pH 8.8 at 25°C), 200 mM (NH4)2SO4, 0.1% (v/v) Tween 20), 1 µL 10 mM dNTP, 1.5 µL 50 mM MgCl2, 1 µL of 10 µM each forward and reverse primer, 0.2 µL Platinum Taq DNA polymerase (Invitrogen) and 36.3 µL nuclease free water (Ambion). For the ITS region, the PCR reaction consisted of an initial denaturation of 94°C for 5 min, followed by 34 cycles of 1 min at 94°C, 30 s annealing at 55°C and 1 min extension at 72°C; and a final extension at 72°C for 10 min. For the amplification of TEF-1α region, the stepdown PCR procedure (Zhang & Gurr, Citation2000) was used in which annealing temperature was reduced from 68°C to 66°C after 5 cycles and then to 62°C after another 5 cycles. PCR reaction consisted of an initial denaturation of 94°C for 5 min, followed by 5 cycles of 1 min denaturation at 94°C, 45 s annealing at 68°C and 1.5 min extension at 72°C, followed by 5 cycles of 1 min denaturation at 94°C, 45 s annealing at 66°C and 1.5 min extension at 72°C, followed by 24 cycles of 1 min denaturation at 94°C, 45 s annealing at 62°C and 1.5 min extension at 72°C; and a final extension at 72°C for 10 min. The PCR products were separated in a 1% (w/v) agarose gel in 1× Tris-borate-EDTA buffer. For sequencing, PCR products were cleaned up using ExoSAP-IT™ PCR Product Cleanup kit (Applied Biosystems) following the manufacturer’s instructions. Sanger Sequencing was performed at McGill University and Genome Quebec Innovation Centre using the Applied Biosystem’s 3730xl DNA Analyzer Technology. The resulting ITS and TEF-1α sequences from forward and reverse primers were aligned using multalin interface (http://multalin.toulouse.inra.fr/multalin/cgi-bin/multalin.pl). The consensus partial sequences of both ITS and TEF-1α were used to retrieve related fungal sequences from the National Centre for Biotechnology Information (NCBI) GenBank database. Of the first 500 BLAST hits for each gene, only 11 were selected that contained both gene sequences of the same isolate. ITS and TEF-1α were combined for these 11 sequences and MEGA7 was used for sequence alignment using Muscle and Maximum likelihood analyses were used for phylogenetic relationship of the sequences (Kumar et al., Citation2016).
Results and discussion
Morphological characterization
A similar fungal colony consistently developed on PDA from isolations made from stem, blossom and leaf tissues of symptomatic plants from the affected alfalfa field. These isolates initially produced olive green colonies with prominent white to grey margins (). The colonies turned dark grey to green after 21 days (). The underside of the 21-day-old colonies turned black to dark green, and production of crystals in agar medium underneath the mycelium mat was observed (Pryor & Michailides, Citation2002). The colonies produced dark, smooth, septate and branched conidiophores (). Conidia were dark with a short tapered beak and had both vertical and transverse septa (). Conidia were generally produced in chains (). The average size of conidia in 7-day-old cultures was 28.7 ± 0.8 (length) × 12.7 ± 0.5 (width) μm. Based on size and shape of conidia and other cultural and morphological characteristics, all isolates were identified as Alternaria alternata (Simmons, Citation1967, Citation1990).
Pathogenicity tests
The pathogenicity of three isolates from the alfalfa field was confirmed in growth chamber experiments, and all produced typical necrotic symptoms on ‘Blue Jay’ and ‘Spredor 4’ alfalfa plants (). The first symptoms started to appear as necrotic spots on leaves 5 days after inoculation. As disease further progressed, necrotic spots enlarged and eventually coalesced, and some infected leaves dried up and fell off (). Necrotic symptoms also developed on stems of the inoculated plants which eventually led to tissue death (). Following the re-isolation from infected leaf sections, the developing fungal colonies on PDA were identical in colony morphology to the original A. alternata isolates from alfalfa. Non-inoculated control plants developed no disease symptoms and were negative for fungal isolation. The results were verified by repeating the experiment and obtaining the same results.
Molecular characterization and phylogenetic analysis
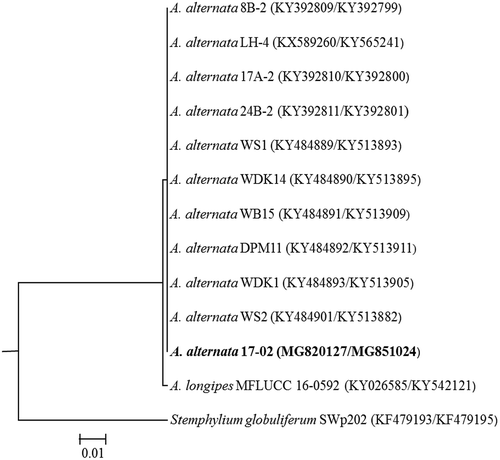
The amplified sequences from both ITS5.8S rDNA (541 bp) and TEF-α (606 bp) of all three A. alternata isolates from alfalfa were identical. Both ITS and TEF-1α sequences of the representative isolate 17–02 were deposited in the NCBI nucleotide database with GenBank accession numbers MG820127 and MG851024, respectively. The combined ITS and TEF-1α (1147 bp) DNA sequences of only isolate 17–02 were used in phylogenetic analysis (Fig. 2). The phylogenetic tree showed that the A. alternata isolate from alfalfa clustered with A. alternata from Aloe vera (Chen & Zhong, Citation2017) and other hosts with 100% similarity (). Alternaria alternata isolate 17–02 was separated from A. longipes causing leaf spot disease in tobacco and other Solanaceous host plants, as well as a closely related alfalfa pathogen, Stemphylium globuliferum, that also causes foliar symptoms and was used as an outgroup (Samac et al., Citation2014).
Fig. 2 Phylogenetic relationship of Alternaria alternata isolate 17–02 from alfalfa with 10 other A. alternata isolates from other hosts together with one isolate of A. longipes and Stemphylium globuliferum. Tree was constructed with combined partial sequences of ITS-5.8S region of rDNA and TEF-1α region. DNA sequences of A. alternata from other hosts and related pathogens were retrieved from NCBI GenBank database and were aligned using Muscle. The isolate from the present study is shown in bold and the GenBank accession numbers are in brackets (for rDNA-ITS/TEF-1α), respectively. Stemphylium globuliferum was used as an outgroup.
Alternaria is a common pathogen found worldwide and it causes distinct symptoms on foliage of different hosts. New disease reports of Alternaria pathogens causing leaf spots and blight symptoms on alfalfa have also been reported from Brazil (Ávila et al., Citation2015), China (Wang & Wang, Citation2010) and India (Maiti et al., Citation2007). The reports from Brazil and China also found A. alternata as the causal agent, whereas the first report from India described the pathogen as A. longipes based on cultural and morphological observations only, but no confirmation was made by molecular methods e.g. sequence analysis.
In conclusion, this is the first report from Canada of a new disease of alfalfa caused by A. alternata. This new disease report should help in recognizing the disease from symptoms on plants in the field, and providing the basis for devising disease management strategies for alfalfa. As most Alternaria diseases usually affect older senescing tissues or stressed plants, it is possible that the insecticide spray used in the spring may have stressed the alfalfa plants, predisposing them to A. alternata infection. Further molecular studies with A. alternata isolates from alfalfa are also needed to define their lineage within A. alternata species group (Armitage et al., Citation2015), and their phylogeny within section Alternaria (Ozkilinc et al., Citation2018).
Acknowledgements
Technical help of Beata Lees and Eric Bevis is gratefully acknowledged. We also thank Zoë Migicovsky for her help in selecting ITS and TEF-1α sequences shared by same isolates from GenBank.
Additional information
Funding
References
- Al-Mughrabi KI, Vikram A, Peters RD, Howard RJ, Grant L, Barasubiye T, Lynch K, Poirier R, Drake KA, Macdonald IK, et al. 2013. Efficacy of Pseudomonas syringae in the management of potato tuber diseases in storage. Biol Control. 64:315–322.
- Armitage AD, Barbara DJ, Harrison RJ, Lane CR, Sreenivasaprasad S, Woodhall JW, Clarkson JP. 2015. Discrete lineages within Alternaria alternata species group: identification using new highly variable loci and support from morphological characters. Fungal Biol. 119:994–1006.
- Ávila MR, Agnol MD, Koshikumo ESM, Martinelli JA, Silva GBP, Schneider-Canny R. 2015. First report in southern Brazil of Alternaria alternata causing Alternaria leaf spot in alfalfa (Medicago sativa). African J Agric Res. 10:491–493.
- Chen Y, Zhong J. 2017. First report of Alternaria alternata causing leaf spot of Aloe vera in China. Plant Dis. 101:1544.
- Cowling WA, Gilchrist DG, Graham JH. 1981. Biotypes of Stemphylium botryosum on alfalfa in North America. Phytopathology. 71:679.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Maiti CK, Sen S, Paul AK, Acharya K. 2007. First report of leaf blight of Medicago sativa caused by Alternaria longipes. J Plant Pathol. 89(Supplement 3):S69.
- Nutter FW Jr, Guan J, Gotlieb AR, Rhodes LH, Grau CR, Sulc RM. 2002. Quantifying alfalfa yield losses caused by foliar diseases in Iowa, Ohio, Wisconsin, and Vermont. Plant Dis. 86:269–277.
- Ozkilinc H, Rotondo F, Pryor BM, Peever TL. 2018. Contrasting species boundaries between sections Alternaria and Porri of the genus Alternaria. Plant Pathol. 67:303–314.
- Pryor BM, Michailides TJ. 2002. Morphological, pathogenic, and molecular characterization of Alternaria isolates associated with Alternaria late blight of pistachio. Phytopathology. 92:406–416.
- Samac DA, Willbur J, Behnken L, Blonde G, Halfman B, Jensen B, Sceaffer C. 2014. First report of Stemphylium globuliferum causing Stemphylium leaf spot on alfalfa (Medicago sativa) in the United States. Plant Dis. 98:993.
- Shahin EA, Shepard JF. 1979. An efficient technique for inducing profuse sporulation of Alternaria species. Phytopathology. 69:618–620.
- Simmons EG. 1967. Typification of Alternaria, Stemphylium, and Ulocladium. Mycologia. 59:67–92.
- Simmons EG. 1990. Alternaria themes and variations (27-53). Mycotaxon. 37:79–119.
- Stuteville DL, Erwin DC. 1990. Compendium of alfalfa diseases. 2nd ed. St. Paul (MN): American Phytopathology Society.
- Wang T, Wang S. 2010. The pathogen of the Medicago sativa leaf spot in Gansu Province. Acta Agrestia Sinica. 18:372–377.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; p. 315–322.
- Zhang Z, Gurr SJ. 2000. Walking into the unknown: a ‘step down’ PCR-based technique leading to the direct sequence analysis of flanking genomic DNA. Gene. 253:145–150.
