Abstract
The genus Verticillium contains 10 plant pathogenic species that are responsible for billions of dollars of crop losses annually on more than 400 plant species worldwide. On alfalfa, Verticillium alfalfae causes Verticillium wilt, a disease with the potential to reduce yields of susceptible cultivars by the second harvest year and to limit the productive stand life to less than 3 years. Susceptible alfalfa cultivars exhibit disease symptoms that include chlorotic V-shaped lesions at the leaf tip that cause entire leaflets to become bleached and twisted, leading to defoliation. Stems often remain green after all leaflets have desiccated, and discolouration, stunting and wilting of shoots is visible. Internally, the taproot can show yellow to brown vascular discolouration. International quarantines sometimes prohibit the entry of alfalfa samples contaminated with the Verticillium pathogen. In this investigation, we developed a recombinase polymerase amplification (RPA) assay coupled with a lateral flow device for the specific and sensitive detection of V. alfalfae. The primers targeted the translation elongation factor 1α (TEF-1α) gene and were designed with mismatches to increase the specificity of the assay. This assay detected as little as 800 fg of DNA from all tested isolates of V. alfalfae. Moreover, non-specific amplification of closely related V. nonalfalfae or other Verticillium species was not observed. Detection of V. alfalfae in inoculated alfalfa cultivars with high or low resistance further demonstrated the specificity and sensitivity of the diagnostic assay. Additionally, RPA detected V. alfalfae from inoculated plants using a crude DNA extraction method. The assay is easy to use and will allow growers, diagnostic labs and regulatory agencies to determine if V. alfalfae is present in alfalfa products.
Résumé
Le genre Verticillium comporte 10 espèces pathogènes qui s’attaquent à plus de 400 plantes commerciales dans le monde et qui sont responsables de milliards de dollars en pertes chaque année. Chez la luzerne, Verticillium alfalfae cause la flétrissure verticilienne, une maladie capable de réduire les rendements des cultivars réceptifs dès la deuxième année de récolte et d’abréger leur vie productive à moins de trois ans. Les cultivars de luzerne réceptifs à l’égard de la maladie affichent des symptômes qui incluent des lésions chlorotiques en V à la pointe des feuilles qui provoquent le blanchiment et la torsion de folioles entières, ce qui entraîne la défoliation. Les tiges demeurent souvent vertes après que toutes les folioles sont desséchées et que la décoloration, le rabougrissement et le flétrissement des pousses sont visibles. À l’intérieur, la racine pivotante peut afficher une décoloration vasculaire variant du jaune au brun. Des quarantaines internationales interdisent parfois l’entrée d’échantillons de luzerne contaminés par l’agent pathogène. Dans le cadre de cette étude, nous avons développé un biotest d’amplification par recombinase-polymérase (ARP) couplé à un dispositif de flux latéral, sensible et propre à la détection de V. alfalfae. Les amorces ciblaient le gène du facteur d’élongation de la traduction (TEF-1α) et ont été conçues avec des mésappariements pour accroître la spécificité du biotest. Ce dernier a pu détecter aussi peu que 800 fg d’ADN, et ce, chez tous les isolats de V. alfalfae testés. En outre, l’amplification non spécifique de V. nonalfalfae, étroitement apparenté, ou d’autres espèces de Verticillium n’a pas été observée. La détection de V. alfalfae dans les cultivars de luzerne inoculés affichant une résistance faible ou élevée a démontré la spécificité et la sensibilité du test diagnostique. De plus, l’ARP a détecté V. alfalfae dans les plants inoculés en utilisant une méthode rudimentaire d’extraction de l’ADN. Le biotest est facile à utiliser et permettra aux producteurs, aux laboratoires de diagnostic et aux organismes de réglementation de déterminer si V. alfalfae a contaminé des produits à base de luzerne.
Introduction
The genus Verticillium consists of a small group of plant pathogenic fungi that cause billions of dollars of damage annually on over 400 plant hosts in many parts of the world (Pegg & Brady, Citation2002). Verticillium species are soilborne and can survive for long periods in soil in the absence of a host as small, melanized resting structures (Wilhelm, Citation1955). These species cause Verticillium wilt, a plant disease that affects the vascular tissues of plants (Pegg & Brady, Citation2002). Host infection starts through the roots and progresses through the vasculature into the shoots (Pegg & Brady, Citation2002).
Verticillium wilt of susceptible alfalfa (Medicago sativa L.) cultivars caused by Verticillium alfalfae Inderb. et al. has the potential to reduce yields as early as the second harvest year and can limit the productive stand life to 3 years or less (Stuteville & Erwin, Citation1990). It was previously one of the most serious diseases on alfalfa in the USA and Canada (Atkinson, Citation1981; Gordon et al., Citation1989), but resistant cultivars have largely eliminated the problem (Stout et al., Citation1998; Huang, Citation2003). Nevertheless, V. alfalfae is considered a quarantine pathogen that could be spread easily to other countries where genetic resistance is not available through contaminated seed or alfalfa hay (Walker, Citation1990; Huang et al., Citation1999; Xu et al., Citation2016). It is believed that the pathogen was introduced into the USA from Europe by contaminated seed (Christen, Citation1982) and it was first reported in Washington State in 1976 (Graham et al., Citation1977). Susceptible alfalfa cultivars exhibit disease symptoms that include v-shaped chlorosis in leaflets, leaves that become pink in colour after desiccation, and discolouration, stunting and wilting of shoots (Stuteville & Erwin, Citation1990). The pathogen is spread in the field primarily during mechanical harvest of hay, but insects can also serve as vectors for the fungus (Harper & Huang, Citation1984). The pathogen overwinters as dark thick-walled mycelium in plant debris, infected plants, and possibly on weed hosts (Stuteville & Erwin, Citation1990; Pegg & Brady, Citation2002), as well as small, melanized resting structures (Wilhelm, Citation1955).
Recent advances have changed our understanding of the genetic diversity in the genus Verticillium. In total, 10 Verticillium species are recognized, including V. dahliae Kleb., V. tricorpus I Isaac, V. nubilum Pethybr., V. longisporum (C. Stark) Karapapa, Bainbr. & Heale, and V. albo-atrum Reinke & Berthold, and five recently described species: V. alfalfae, V. nonalfalfae Inderb. et al., V. zaregamsianum Inderb. et al., V. isaacii Inderb. et al. and V. klebahnii Inderb. et al. (Inderbitzin et al., Citation2011). Verticillium alfalfae was discovered as a distinct lineage within V. albo-atrum, leading to its description as a new species. Inderbitzin et al. (Citation2013) later developed a diagnostic PCR assay for the specific detection of V. alfalfae, as well as the other Verticillium species. This assay is the only one available that can differentiate V. alfalfae from the closely related V. nonalfalfae. The accurate detection of V. alfalfae as the causal-agent of verticillium wilt in alfalfa is particularly important not only because of the potential yield reduction (up to 50%) in susceptible cultivars (Peaden et al., Citation1985), but also because of quarantines imposed by certain countries to prevent the entry of alfalfa and hay containing V. alfalfae (Walker, Citation1990; Huang et al., Citation1999).
The use of recombinase polymerase amplification (RPA) (TwistDx Ltd, Cambridge, UK) allows for the rapid, easy, reproducible, sensitive and specific detection of pathogens in laboratory and field settings by experienced and inexperienced individuals. RPA uses isothermal technology similar to loop-mediated-isothermal amplification (LAMP) and helicase-dependent amplification (HDA) (Biohelix Corp, Beverly, MA), but with clear advantages. The design of the primers and probes is easier for RPA assays compared with LAMP assays, and RPA technology is not proprietary like HDA, which means that anyone can develop their own detection assays for research purposes (Vincent et al., Citation2004; Miles et al., Citation2015). The kits come with a lyophilized pellet containing reagents and all that needs to be added by the user are primers, probe, rehydration buffer (supplied by manufacturer) and 1 µL of a sample that can be pure genomic DNA or crude DNA extract. To start the amplification, magnesium acetate is added to the reaction and the tube is placed at 37–40°C for 20 min. Detection of amplification can be accomplished in three different ways: (i) agarose gel electrophoresis as performed in end-point PCR (TwistDx – TwistAmp Basic or Agdia – AmplifyRP); (ii) fluorescence as in real-time PCR (TwistDx – TwistAmp exo or Agdia – AmplifyRP XRT Discovery kit); or (iii) coupled with a lateral flow device like a ‘sandwich-assay’ using a lateral-flow strip (TwistDx – TwistAmp nfo or Agdia – AmplifyRP Acceler8 Discovery kit).
The objectives of this investigation were to: (i) develop a RPA assay coupled with a lateral-flow device for the specific and sensitive detection of V. alfalfae; (ii) identify a crude tissue extraction protocol for RPA detection of V. alfalfae in planta; and (iii) develop a rapid, field-deployable diagnostic assay that can be used directly at the point of sample collection.
Materials and methods
Verticillium isolates
In total, 34 Verticillium isolates were used in this study representing the 10 species that comprise the genus (). These isolates included 17 V. alfalfae (including isolates originally labelled as V. albo-atrum), one V. nonalfalfae, one V. albo-atrum, three V. dahliae, two V. isaacii, two V. klebahnii, one V. nubilum, one V. tricorpus, one V. zaregamsianum and five V. longisporum (including three A1/D1 and two A1/D2) isolates. The identities of the 10 species were confirmed using the primers developed by Inderbitzin et al. (Citation2013) for end-point PCR. The cultures were maintained on half-strength potato dextrose agar (½ PDA) at room temperature. To produce hyphal mats, four 0.25 mm2 plugs were cut from cultures grown on ½ PDA and transferred to 250 mL of potato dextrose broth (PDB). The PDB cultures were maintained at room temperature (20–22°C) in complete darkness, and the hyphae were collected after 15–17 days. The hyphae were dried in sterile paper towels for 24 h and ground using a sterile mortar and pestle.
Table 1. Detection or lack of detection of Verticillium species isolates by the V. alfalfae-specific RPA assay. The RPA assay detected all V. alfalfae isolates. Note that isolates previously described as V. albo-atrum on alfalfa were found to be V. alfalfae based on species-specific PCR assays developed by Inderbitzin et al. (Citation2013).
Marker design for RPA and assay development
A partial region of the translocation elongation factor 1 (TEF-1α) gene was chosen as the target for the detection assay because of its conserved nature between V. alfalfae isolates and for the number of polymorphisms when compared with the other Verticillium species. Sequence alignments of partial sequences of the TEF-1α gene used for the target were conducted using the program Geneious (v.9.1.5; Biomatters Ltd, Auckland, New Zealand) and included all 10 Verticillium species. The primers and probe were manually designed based on alignments. The specificity of the sequences was further confirmed in silico by using BLASTn (Altschul et al., Citation1990). The selected amplicon had a length of 219 nucleotides.
Ten forward and 10 reverse primers were designed (between 27 and 36 bp) to test various combinations and lengths. A forward primer with 8 mismatches (Valf-TEF-F8m) and a reverse primer with 9 mismatches (Valf-TEF-R9m) were chosen for the greatest specificity (). A reverse primer with 12 mismatches (Valf-TEF-R12m) was also tested, but it had lower sensitivity (). The mismatches were added because the original primers designed showed non-specific amplification of the sister species V. nonalfalfae and more differences in the sequences were necessary to prevent the amplification and detection of non-specific amplicons. Previous research showed that adding mismatches to primers used in RPA assays did not significantly affect detection; it decreases the sensitivity but it has the potential of increasing the specificity (Boyle et al., Citation2013; Daher et al., Citation2015). The primers and probe were designed following the recommendations of the TwistAmp nfo kit manufacturer (TwistDx Ltd, Cambridge, UK), and were ordered from Integrated DNA Technologies – IDT, Inc. and Biosearch Technologies, Inc. (Petaluma, CA) respectively. The reverse primers were labelled with the biotin antigen at their 5ʹ end; and the probe was designed with a carboxyfluorescein group (FAM) at its 5ʹ end, a tetrahydrofuran (THF also known as dSpacer) replacing a nucleotide 30 nucleotides downstream from the 5ʹ end, and a polymerase extension blocking group (Spacer C3) at the 3ʹ end.
Table 2. Forward and reverse primers (with mismatches underlined), and probe used in RPA assay. The assay’s sensitivity decreased without affecting the detection in inoculated alfalfa plants, and the specificity increased.
The RPA reaction was performed per the manufacturer’s directions (TwistDx, Cambridge, UK). The assay was conducted at 37°C for 20 min in a 10 µL reaction composed of 5.9 µL rehydration buffer, 0.42 µL forward primer (10 µM), 0.42 µL reverse primer (10 µM), 0.12 µL probe (10 µM) 0.5 µL magnesium acetate (280 mM), 1.64 µL molecular grade water and 1 µL DNA template.
Exclusivity and inclusivity panels of RPA marker
Isolates representing the 10 species of Verticillium were tested in exclusivity and inclusivity panels to check the specificity of the assay. A published end-point PCR assay (Inderbitzin et al., Citation2013) was used to confirm the identity of the isolates and the results of the developed RPA assay () because of its high specificity to differentiate the 10 different Verticillium species. Moreover, sensitivity assays were tested with concentrations ranging from 1 ng µL−1 to 1 fg µL−1 in 10-fold dilutions of DNA from 17 different V. alfalfae isolates. Initial concentrations were measured using the Qubit dsDNA BR assay kit (Thermo Fischer Scientific, Waltham, MA) to ensure proper quantification of initial DNA concentrations of V. alfalfae.
RPA validation in inoculated plants and crude DNA extraction
Seeds of alfalfa cultivars with high resistance (55V50) and low resistance (‘La Jolla’) to Verticillium wilt were sown in 72-cell flats (25 cm × 50 cm) containing soilless potting mix (Sunshine LA4 PC). Four weeks after sowing, 10 plants of each cultivar were unpotted and the roots were rinsed with distilled water to remove the potting mix. Five plants of each cultivar were inoculated with V. alfalfae isolate PD353 () by trimming off ~1.5 cm of the root tips and then submerging the roots in a slurry containing 1 g of ground hyphae in 10 mL of 0.01% agarose for 30 s before repotting (Gaige et al., Citation2010). Five negative control plants of each cultivar were treated the same way, but were inoculated with sterile 0.01% agarose. The plants were grown in a growth chamber with a 14-h photoperiod set at 21°C day/16°C night in a completely randomized design.
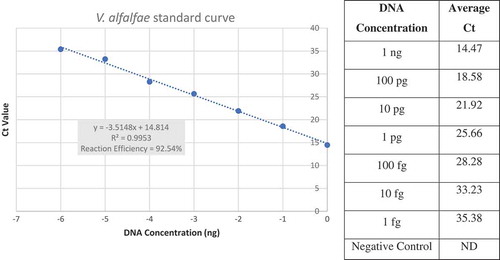
Four weeks after inoculation, the plants were removed from the potting mix and the entire shoot (leaf and stem tissue) of each plant was excised at the crown using sterile scissors. DNA was extracted from each shoot using the MP FastDNA Spin Kit (MP Biomedicals, Santa Ana, CA) per the manufacturer’s instructions. DNA extractions were quantified using the Qubit dsDNA BR kit and adjusted to 10 ng µL−1 for use in subsequent end-point PCR, quantitative PCR (qPCR) and RPA assays to detect the pathogen. RPA was conducted as described in the manufacturer’s instructions. Infection by V. alfalfae was further confirmed using a published end-point PCR protocol (Inderbitzin et al., Citation2013) and an unpublished qPCR assay using the rDNA intergenic region (Bilodeau et al., Citation2012; G. Bilodeau, personal communication). This unpublished qPCR assay reliably distinguished V. alfalfae from the other Verticillium species in preliminary testing, and was used instead of the qPCR assay developed by Larsen et al. (Citation2007), which was developed before the taxonomic separation of V. alfalfae from V. albo-atrum (Inderbitzin et al., Citation2011). Additionally, the qPCR protocol was used to build a standard curve to relate Ct value to DNA concentration. Verticillium alfalfaegenomic DNA was diluted 10-fold and tested from 1 ng to 1 fg per mixture. Initial DNA concentrations were determined using a Qubit dsDNA BR kit. Each reaction was performed in three replicates and the average data were used to build a standard curve with Ct in the y-axis and amount of DNA (in ng) in the x-axis, using a linear fit. The equation derived from the standard curve provides information about the amount of V. alfalfae present in the inoculated plants that can be used with the qPCR test results.
A crude DNA extraction was also tested from inoculated and negative control plants to determine if the RPA assay could be used under field conditions where DNA extraction kits are not practical. Crude DNA extraction was performed using the protocols of Satya et al. (Citation2013) and Londoño et al. (Citation2016) with minor modifications. Sodium hydroxide (0.5 N) was used to homogenize fresh plant tissue in a plastic bag, using 10 µL of buffer per mg of plant tissue. Crude extracts were centrifuged at 10,000 g for 15 s to precipitate large particles and the supernatant was used as the template.
Finally, the RPA assay also was tested with alfalfa plant and soil samples that were collected from six alfalfa fields in Central Oregon (Jefferson County) in September 2016. All six fields were apparently healthy, but efforts were made to find plants that appeared stunted, chlorotic, or wilted. However, the widespread use of V. alfalfae-resistant cultivars since the 1990s made it impossible to find fields with classical symptoms of Verticillium wilt and historical disease records about the specific fields sampled were not available. Previous surveys found V. alfalfae throughout large areas of the Columbia Basin, which includes Jefferson County (Arny & Grau, Citation1985). Therefore, the RPA assay was used to determine if V. alfalfae was present and potentially causing asymptomatic infection in resistant alfalfa cultivars, which would result in export restrictions. Two plant and two soil samples were collected from different locations in each field. Samples from each field were then pooled together and DNA was extracted from both soil and root samples using the MP FastDNA Spin Kit as described above. The results of the RPA test were validated with the unpublished qPCR assay.
Results and discussion
In this study, we developed an RPA assay coupled with a lateral-flow device that uses DNA crude extract samples (0.5 N NaOH). This makes the V. alfalfae-specific RPA assay a rapid, field-deployable diagnostic tool that can be used both in the field and lab. The RPA assay detected V. alfalfae directly from pure cultures and from inoculated plants using a commercial kit for DNA extraction and a crude DNA extraction method (, , ). The assay detected at least 800 fg µL−1 of DNA from pure cultures of the V. alfalfae isolates when primers Valf-TEF-F8m and Valf-TEF-R9m were used (). Results from the end-point PCR and the qPCR assay confirmed the presence of V. alfalfae and demonstrated the efficiency and robustness of the RPA assay.
Fig. 1 (Colour online) The RPA assay did not cross-react with any of the other Verticillium species.

Fig. 2 (Colour online) The RPA assay had a consistent sensitivity of at least 800 fg µL−1 DNA in all tested V. alfalfae isolates.

Mismatches in the forward and reverse primers () did not prevent detection of V. alfalfae and increased the specificity of the assay, while primers without the mismatches also amplified V. nonalfalfae DNA. The mismatches were robust for all assays, meaning that the primers amplified all V. alfalfae isolates tested and no other Verticillium species. Although the sensitivity of the RPA assay was reduced from 10 fg µL−1 to 800 fg µL−1 due to the mismatches in the primers, the assay was still sensitive enough to detect V. alfalfae in all inoculated alfalfa plants regardless of the level of resistance to Verticillium wilt (see below). When the assay was performed on V. alfalfae DNA using primer Valf-TEF-R12m with 12 mismatches (), the sensitivity of the assay decreased to 1 pg µL−1 (data not shown) compared with the greater sensitivity (800 fg µL−1) observed when using primer Valf-TEF-R9m with nine mismatches was used (). Therefore, all subsequent assays were performed using Valf-TEF-R9m. The qPCR data were used to build a standard curve which had a high correlation coefficient (R2 = 0.9953) and a reaction efficiency of 92.54% calculated with the equation E = [(10–1/slope – 1) *100] (). These values indicated that the assay results were accurate across a seven-fold dilution of DNA and amplification of the target was not inhibited.
Fig. 3 (Colour online) Verticillium alfalfae standard curve for qPCR demonstrated specificity and sensitivity of the assay. On the x-axis DNA concentrations were logarithmically transformed to obtain a linear graph. On the y-axis, the calculated Ct value is shown. Correlation and reaction efficiency values demonstrate that the reaction is accurate and amplification was not inhibited.
Verticillium alfalfae was detected in all inoculated plants (cultivars 55V50 and ‘La Jolla’) by RPA () and by qPCR (). End-point PCR was only able to detect the presence of V. alfalfae in two inoculated plants of cultivar ‘La Jolla’ (, ).
Table 3. Detection of V. alfalfae in highly resistant (HR) and low resistant (LR) alfalfa cultivars that were artificially inoculated using the RPA assay. Confirmation assays were performed using end-point PCR and qPCR.
Fig. 4 (Colour online) The RPA assay detected V. alfalfae in high resistance (55V50) and low resistance (‘La Jolla’) artificially inoculated alfalfa plants.
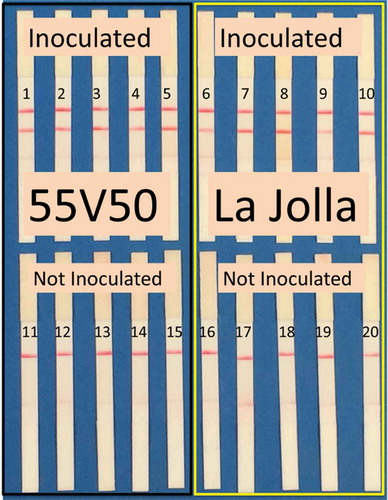
Fig. 5 (Colour online) End-point PCR detected V. alfalfae in only two of the inoculated plants of the low resistance cultivar ‘La Jolla’ (samples 6 and 7) using Inderbitzin et al. (Citation2013) with diagnostic bands with a size of 1060 bp. On the other hand, RPA and qPCR confirmed the presence of V. alfalfae in all inoculated plants. The molecular ladder was AMRESCO 100bp E-Z vision ladder.

RPA detected at least 3.25 pg of V. alfalfae DNA in infected alfalfa plants (). Previous experiments showed similar results in detecting V. alfalfae in inoculated plants with a real-time PCR assay (Larsen et al., Citation2007). However, V. alfalfae was not detected from any of the soil or plant field samples that were collected from central Oregon and tested using the RPA assay (data not shown), and the lack of detection was validated by obtaining the same results using the qPCR assay. Given the pathogen’s widespread presence in this region previously (Arny & Grau, Citation1985), this may suggest that V. alfalfae does not persist in fields where resistant alfalfa cultivars are used. However, further research is needed on V. alfalfae persistence in fields with a known past history of verticillium wilt and then planted with resistant cultivars before a definitive conclusion can be made. Additional research is also needed to confirm whether V. alfalfae is still present in other alfalfa fields regionally and to clarify the impact of resistant cultivars on seed contamination.
Finally, all inoculated alfalfa plants produced positive RPA results with the crude DNA extraction using 0.5 N NaOH (using 10 µL of buffer per mg of plant tissue) using primers Valf-TEF-F8m and Valf-TEF-R9m. This greatly decreased the processing time for the samples and allowed for its use directly in the field. The assay is easy-to-use and will allow growers, diagnostic labs and regulatory agencies to determine whether V. alfalfae is present in alfalfa products, including seed and hay.
Acknowledgements
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture (USDA). The authors thank Debby Samac, Krishna Subbarao and Patrik Inderbitzin for providing Verticillium isolates and Guillaume Bilodeau for sharing V. alfalfae-specific qPCR procedures.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Arny DC, Grau CR. 1985. Importance of verticillium wilt of alfalfa in North America. Can J Plant Pathol. 7:187–190.
- Atkinson TG. 1981. Verticillium wilt of alfalfa: challenge and opportunity. Can J Plant Pathol. 3:266–272.
- Bilodeau GJ, Tropiano R, Briere SC. 2012. Development of diagnostic assays for detection of Verticillium in alfalfa and flax and detection of blackleg (Leptosphaeria maculans) in canola using real-time PCR [abstract]. Phytopathology. 102:S4.12.
- Boyle DS, Lehman DA, Lillis L, Peterson D, Singhal M, Armes N, Parker M, Piepenburg O, Overbaugh J. 2013. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. MBio. 4:e00135–13.
- Christen AA. 1982. Demonstration of Verticillium albo-atrum within alfalfa seed. Phytopathology. 72:412–414.
- Daher RK, Stewart G, Boissinot M, Boudreau DK, Bergeron MG. 2015. Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol Cell Probe. 29:116–121.
- Gaige AR, Ayella A, Shuai B. 2010. Methyl jasmonate and ethylene induce partial resistance in Medicago truncatula against the charcoal rot pathogen Macrophomina phaseolina. Physiol Mol Plant Pathol. 74:412–418.
- Gordon TR, Correll JC, Gilchrist DG, Martensen AN. 1989. Verticillium wilt of alfalfa in California. Plant Dis. 73:18–20.
- Graham JH, Peaden RN, Evans DW. 1977. Verticillium wilt of alfalfa found in the United States. Plant Dis Rep. 61:337–340.
- Harper AM, Huang HC. 1984. Contamination of insects by the plant pathogen Verticillium albo-atrum in an alfalfa field. Environ Entomol. 13:117–120.
- Huang CH, Acharya SN, Hou TJ, Erickson RS, Dalton RE, Mueller CA. 1999. Susceptibility of Chinese alfalfa cultivars to verticillium wilt. Plant Pathol Bull. 8:67–72.
- Huang HC. 2003. Verticillium wilt of alfalfa: epidemiology and control strategies. Can J Plant Pathol. 25:328–338.
- Inderbitzin P, Bostock RM, Davis RM, Usami T, Platt HW, Subbarao KV. 2011. Phylogenetics and taxonomy of the fungal vascular wilt pathogen Verticillium, with the descriptions of five new species. PloS One. 6:e28341.
- Inderbitzin P, Davis RM, Bostock RM, Subbarao KV. 2013. Identification and differentiation of Verticillium species and V. longisporum lineages by simplex and multiplex PCR assays. PloS One. 8:e65990.
- Larsen RC, Vandemark GJ, Hughes TJ, Grau CR. 2007. Development of a real-time polymerase chain reaction assay for quantifying Verticillium albo-atrum DNA in resistant and susceptible alfalfa. Phytopathology. 97:1519–1525.
- Londoño MA, Harmon CL, Polston JE. 2016. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol J. 13:48.
- Miles TD, Martin FN, Coffey MD. 2015. Development of rapid isothermal amplification assays for detection of Phytophthora spp. in plant tissue. Phytopathology. 105:265–278.
- Peaden RN, Gilbert RG, Christen AA. 1985. Control of Verticillium albo-atrum on alfalfa. Can J Plant Pathol. 7:511–514.
- Pegg GF, Brady BL. 2002. Verticillium wilts. Oxfordshire (UK): CABI Publishing.
- Satya P, Mitra S, Ray DP, Mahapatra BS, Karan M, Jana S, Sharma AK. 2013. Rapid and inexpensive NaOH based direct PCR for amplification of nuclear and organelle DNA from ramie (Boehmeria nivea), a bast fibre crop containing complex polysaccharides. Indian Crop Prod. 50:532–536.
- Stout DG, Acharya SN, Hall JW, Broersma K. 1998. Yield and persistence from mixtures of a winter hardy and a Verticillium wilt resistant lucerne cultivar. J Agron Crop Sci. 180:27–31.
- Stuteville DL, Erwin DC. 1990. Compendium of alfalfa diseases. St Paul (MN): American Phytopathological Society.
- Vincent M, Xu Y, Kong H. 2004. Helicase‐dependent isothermal DNA amplification. EMBO Rep. 5:795–800.
- Walker J. 1990. Verticillium albo-atrum in Australia: a case study of information confusion in plant pathology. Australas Plant Path. 19:57–67.
- Wilhelm S. 1955. Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology. 45:180–181.
- Xu S, Li YZ, Nan ZB. 2016. First report of verticillium wilt of alfalfa caused by Verticillium alfalfae in China. Plant Dis. 100:220.
