Abstract
The whitefly-transmitted begomoviruses (family Geminiviridae) infect dicotyledonous plants and occur in tropical and sub-tropical regions. Although most commonly found in herbaceous plants, recently begomoviruses have increasingly been identified in woody plants. Leaf samples from five mulberry (Morus alba L.) plants with leaf yellowing and curling symptoms were collected in Lahore (Pakistan) and shown by PCR to be associated with a begomovirus, an alphasatellite and a betasatellite. The complete sequences of two begomovirus clones, as well as an alphasatellite clone and a betasatellite clone, were determined. The begomovirus clones were shown to be isolates of ageratum enation virus (AEV), a virus most commonly identified in weeds but increasingly being identified in crops such as tomato, soybean and fenugreek. Analyses of the sequences of the alphasatellite and betasatellite clones showed them to be isolates of Guar leaf curl alphasatellite and Papaya leaf curl betasatellite, respectively. Both the virus and the alphasatellite sequences showed evidence of a recombination. This is the first report of the weed-infecting monopartite begomovirus AEV, and associated satellites, infecting the woody plant mulberry.
Résumé
Les bégomovirus (famille des Geminiviridae) transmis par l’aleurode infectent les dicotylédones des régions tropicales et subtropicales. Bien qu’on les trouve généralement chez les plantes herbacées, récemment, on en détecte de plus en plus chez les plantes ligneuses. Des échantillons de feuilles, provenant de cinq mûriers (Morus alba L.) dont les feuilles affichaient des symptômes de jaunissement et d’enroulement, ont été collectés à Lahore (Pakistan) et, après analyse par PCR, ont été associés à un bégomovirus, à un α-satellite et à un ß-satellite. Les séquences complètes de deux clones de bégomovirus, de même que d’un clone d’α-satellite et d’un de ß-satellite, ont été établies. Les clones de bégomovirus se sont révélé des isolats d’ageratum enation virus (AEV), un virus très souvent associé aux adventices, mais trouvé de plus en plus souvent dans des cultures comme la tomate, le soja et le fenugrec. Les analyses des séquences des clones de l’α-satellite et du ß-satellite ont permis de les identifier en tant qu’isolats de l’α-satellite de la cloque du guar et du ß-satellite de la cloque de la papaye, respectivement. Les séquences du virus et de l’α-satellite ont affiché un certain taux de recombinaison. Il s’agit de la première mention du bégomovirus monopartite AEV, et de ses satellites associés, qui infecte habituellement les adventices herbacées, s’attaquant au mûrier, une plante ligneuse.
Introduction
Viruses belonging to the family Geminiviridae are important phytopathogens which have a characteristic twinned (geminate) morphology and circular single-stranded (css) DNA genomes. This family has recently been extended to nine genera – Becurtovirus, Begomovirus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocovirus and Turncurotovirus (Zerbini et al., Citation2017). The genus Begomovirus is the largest, and includes some of the most widespread and destructive geminiviruses that are transmitted by the whitefly Bemisia tabaci. Begomoviruses have either bipartite genomes with components known as DNA-A and DNA-B of approximately equal size (~ 2.8 kb) or monopartite genomes homologous to the DNA-A components of bipartite begomoviruses (Hanley-Bowdoin et al., Citation2013). The genomes of monopartite begomoviruses and DNA-A components of bipartite begomoviruses encode six conserved open reading frames (ORFs) transcribed from a single bi-directional promotor present in an intergenic region (IR) (Ashraf et al., Citation2014). The products encoded in the virion-sense are involved in virus movement in plants and transmission between plants, whereas those encoded in the complementary-sense are involved in virus replication and control of virus and host gene expression. The DNA-B components of bipartite begomoviruses encode two ORFs, one in each orientation, that are involved in inter- and intracellular virus movement in plants (Rojas et al., Citation2005).
A majority of monopartite begomoviruses associate with additional small (~1.4 kb) cssDNA molecules known as alphasatellites and betasatellites. Alphasatellites are autonomously replicating but require a helper virus for movement in plants and transmission between plants (Mar et al., Citation2017). They are widespread in the Old World (OW) and have most commonly been identified in association with monopartite begomoviruses that are associated with betasatellites (Mansoor et al., Citation1999). However, some alphasatellites have recently also been identified in the New World (NW) in association with bipartite begomoviruses (Paprotka et al., Citation2010; Romay et al., Citation2010). In contrast, betasatellites have so far only been identified in the OW. They encode a single ORF in the complementary sense, the product of which is a pathogenicity determinant and overcomes host plant defences (Eini, Citation2017). Betasatellites are true satellites that require a helper virus not only for movement in plants and transmission between plants but also for initiation of replication (Briddon & Mansoor, Citation2008).
Huge economic losses to crops and ornamental plants are caused by begomoviruses worldwide. Recently, begomoviruses have also been reported to infect a few woody trees, including citrus (Loconsole et al., Citation2012) and mulberry (Ma et al., Citation2015). Mulberry (Morus alba L.) is a woody dicotyledonous plant domesticated for sericulture in temperate, tropical and subtropical regions (Dai et al., Citation2015) and has been reported to be infected by nepoviruses (Tsuchizaki et al., Citation1971) and carlaviruses (Kuai, Citation2010). Recently, an as yet taxonomically unassigned monopartite geminivirus, mulberry mosaic dwarf associated virus (MMDaV), has also been isolated from mulberry plants exhibiting leaf crinkling, mosaic and dwarfing symptoms in China (Lu et al., Citation2015; Ma et al., Citation2015). The study described here shows that mulberry plants with leaf yellowing and curling symptoms in Pakistan are associated with a monopartite begomovirus and both an alphasatellite and a betasatellite.
Materials and methods
Sample collection and isolation of total genomic DNA
Leaf samples of five mulberry plants showing typical symptoms of begomovirus infection, including leaf curling and yellowing (), as well as three non-symptomatic plants () were collected in Lahore, Pakistan (31.29°N, 74.17°E), in December 2013. Total genomic DNA was extracted from leaf tissue using the CTAB method (Doyle & Doyle, Citation1990).
Rolling circle amplification (RCA), cloning and sequencing
Circular DNA molecules in DNA samples were enriched by rolling circle amplification (RCA) using phi29 DNA-polymerase (ThermoScientific) as described earlier (Qurashi et al., Citation2017). The samples were initially analysed for infection by a begomovirus by PCR with degenerate primer pair AC1048/AV494 using diluted RCA product as template (Wyatt & Brown, Citation1996), which amplify the core of the coat protein (CP) gene of begomoviruses. The gel-purified PCR products (~0.6 kb) from all five samples were cloned in pTZ57R/T using a CloneJet PCR cloning kit (ThermoScientific) and selected clones were sequenced (First BASE Laboratories, Sdn Bhd, Malaysia). An abutting primer pair (52-F 5ʹ-GACGAGAATATTAAGACCAAG-3ʹ/52-R 5ʹ-CATCCATATCTTACCCAAGAC-3ʹ) was designed using the resultant CP sequence to amplify the complete begomovirus genome. The universal primer pairs DNA101/DNA102 (Bull et al., Citation2003) and Beta01/Beta02 (Briddon et al., Citation2002) were used to PCR-amplify complete alpha- and betasatellite molecules, respectively. Amplicons were cloned and sequenced in their entirety as described above.
Sequence comparisons and phylogenetic analysis
Sequences were initially analysed using BLASTn in NCBI (https://www.ncbi.nlm.nih.gov). Subsequently, the Species Demarcation Tool (SDT) (Muhire et al., Citation2014) was used to determine pair-wise percentage nucleotide (nt) sequence identities of all the clones to the respective sequences retrieved from GenBank. Evolutionary relationships were determined by constructing phylogenetic dendrograms using the neighbour-joining algorithm available in MEGA7 software (Tamura et al., Citation2013). ORFs in sequences were identified using the NCBI ORF finder tool (http://www.ncbi.nlm.nih.gov).
Recombination analysis
Putative recombination events were identified using seven different algorithms available in the Recombination Detection Program (RDP4) (Martin et al., Citation2015).
Results and discussion
Analyses of begomovirus sequences
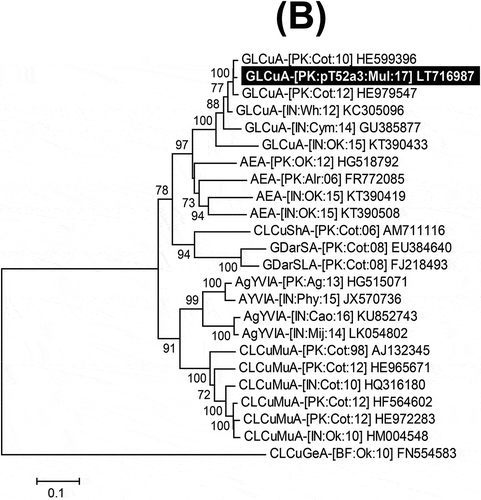
The five symptomatic leaf samples produced amplicons of the expected size whereas no amplicons were produced for samples from non-symptomatic plants. An initial analysis of the core CP sequences from the five symptomatic plants (M51 to M55) showed that all were infected with a begomovirus (results not shown). The complete sequences of two apparently full-length begomovirus clones, pT52-A and pT53-A, were determined from two randomly selected samples (M52 and M53, respectively) and each consisted of 2754 nt. The sequences were submitted to GenBank under accession numbers LT716984, LT716985, respectively. The predicted genome organization of the begomovirus isolates is typical of OW begomoviruses, containing six conserved ORFs (). The two sequences showed 96% nt sequence identity and the highest nt sequence identities (95.5 and 98%, respectively) with an Ageratum enation virus (AEV) variant (JX436473) isolated from fenugreek in India (). Based on the currently applicable species demarcation criteria of the International Committee for the Taxonomy of Viruses (ICTV), for which isolates of two species share less than 91% nt sequence identity (Brown et al., Citation2015), pT52-A and pT53-A are variants of AEV. This conclusion was supported by the phylogenetic analysis in which the sequences from mulberry grouped into a well-supported clade with other isolates of AEV ().
Table 1. Pairwise per cent (%) nucleotide sequence identities of the isolated full-length genomes of ageratum enation virus (AEV) isolates (pT52-A and pT53-A) and their individual ORFs from Morus alba L. to previously reported begomoviruses.
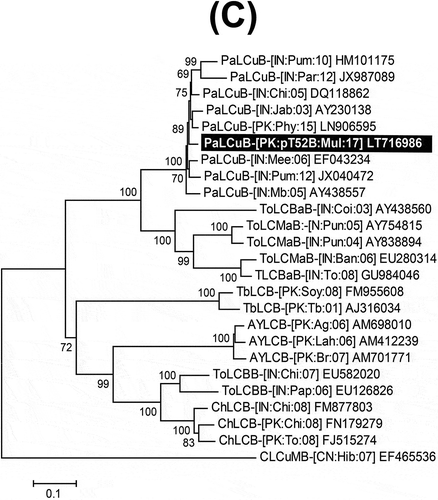
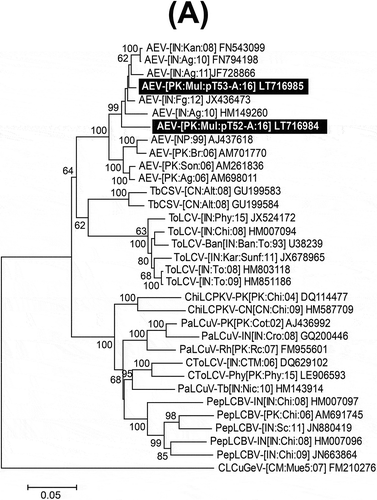
Fig. 2 Neighbour-joining (NJ) phylogenetic dendrograms based on alignments of the complete nucleotide sequences of (A) selected begomovirus; (B) alphasatellite; and (C) betasatellite isolates, using Kimura-2 parameter model available in MEGA7. The virus and satellite isolates from mulberry are highlighted as white text on a black background. Horizontal lines are proportional to nucleotide substitutions per site. Per cent bootstrap values (1000 replicates) are given at branch nodes. Only bootstrap scores above 60% are shown. All isolates used for comparison are represented by their respective accession numbers. Begomovirus, beta- and alphasatellite acronyms and isolate descriptors are according to Brown et al. (Citation2015), Briddon et al. (Citation2008) and Mubin et al. (Citation2009) respectively.
Analyses of the satellite sequences
Putatively full-length alphasatellite (pT52-a3) and betasatellite (pT52B) clones were obtained from sample M52 and the sequences were deposited in GenBank under accession numbers LT716987 and LT716986, respectively. The clone pT52-a3 comprised of 1362 nt showed 98.6% nt sequence identity with an isolate of Guar leaf curl alphasatellite (GLCA; HE599396) isolated from cotton in Pakistan. Phylogenetic analysis showed this sequence to segregate in a well-supported clade with other GLCA isolates (). Based on the suggested species demarcation criteria of >83% for alphasatellites (Mubin et al., Citation2009), pT52-a3 is a variant of GLCA. The sequence of pT52B was comprised of 1356 nt and showed the highest nt sequence identity (97.1%) with Papaya leaf curl betasatellite (PaLCuB; EF043234), isolated from potato in India. Based on the suggested species demarcation criteria for betasatellites of >79% (Briddon et al., Citation2008) pT52B is an isolate of PaLCuB. This conclusion was supported by a phylogenetic analysis showing pT52B to segregate in a well-supported clade with previously reported PaLCuB isolates ().
Analyses of begomovirus and DNA satellites sequences for recombination
The incongruity between sequence identities for the ORFs of pT52-A and pT53-A suggested that these virus isolates might have a recombinant origin (). This hypothesis was supported by recombination analysis using the seven algorithms available in the RDP4 program (Supplementary Data). This predicted three recombination events for pT52-A at nt coordinates 677–1450, 2108–2313 and 2245–2649 with the lowest P-values calculated as 8.94 × 10−13–5.83 × 10−21, 8.18 × 10−04–3.11 × 10−07 and 1.31 × 10−12–3.70 × 10−31, respectively. For pT53-A, two putative recombination events were predicted at nt coordinates 2108–2313 and 2094–2693 with the lowest P-values calculated as 8.18 × 10−04–3.11 × 10−07 and 1.31 × 10−12–3.70 × 10−31, respectively (Supplementary Data). Analysis of pT52-a3 identified one likely recombination event at nt coordinates 717–1171 supported with P-values of 1.22 × 10−04–3.54 × 10−08 (Supplementary Data), while no recombination events were detected in pT52B.
The recent identification of geminiviruses infecting woody plants that include citrus (Loconsole et al., Citation2012), grapevine (Krenz et al., Citation2012), Jatropha (Polston et al., Citation2014) and mulberry (Lu et al., Citation2015; Ma et al., Citation2015) raises concerns that these viruses may increasingly be spreading into woody plants. We confirmed the weed-infecting monopartite begomovirus AEV (Tahir et al., Citation2015), and associated satellites, to be present in mulberry. Furthermore, the identification of a begomovirus infecting mulberry in India, based on partial sequences that are related to Indian cassava mosaic virus (ICMV) and Sri Lankan cassava mosaic virus (SLCMV) (two viruses that infect the woody plant cassava) point towards the growing threat of begomoviruses to woody plants on the Indo-Pak subcontinent (Shery, Citation2016). Further studies are required to assess the extent of begomovirus infection of woody plants, the losses they cause and the threat they pose to, for example, fruit trees.
Supplemental Material
Download MS Word (153.5 KB)Acknowledgements
This research work was funded by a short research grant from University of the Punjab, Lahore, Pakistan. FQ carried out this research work as part of her PhD studies. The authors are thankful to Rob W. Briddon for critically reading and improving the manuscript.
Supplementary material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2018.1490929.
Additional information
Funding
References
- Ashraf MA, Shahid AA, Rao AQ, Bajwa KS, Husnain T. 2014. Functional characterization of a bidirectional plant promoter from Cotton leaf curl Burewala virus using an Agrobacterium-mediated transient assay. Viruses. 6:223–242.
- Briddon RW, Brown JK, Stanley J, Zerbini M, Zhou X, Fauquet CM. 2008. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch Virol. 153:763–781.
- Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. 2002. Universal primers for the PCR-mediated amplification of DNA β: a molecule associated with some monopartite begomoviruses. Mol Biotechnol. 20:315–318.
- Briddon RW, Mansoor S. 2008. Beta ssDNA satellites. In: Mahy BWJ, van Regenmortel MHV, editors. Encyclopedia of virology. Oxford: Academic Press; p. 314–321.
- Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JF, Fiallo-Olivé E, Briddon RW, Hernández-Zepeda C, Idris A, et al. 2015. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol. 160:1593–1619.
- Bull SE, Briddon RW, Markham PG. 2003. Universal primers for the PCR-mediated amplification of DNA 1: a satellite-like molecule associated with begomovirus-DNA β complexes. Mol Biotechnol. 23:83–86.
- Dai F, Wang Z, Luo G, Tang C. 2015. Phenotypic and transcriptomic analyses of autotetraploid and diploid mulberry (Morus alba L.). Inter J Mol Sci. 16:22938–22956.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Eini O. 2017. A betasatellite-encoded protein regulates key components of gene silencing system in plants. Mol Biol. 51:579–585.
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 11:777–788.
- Krenz B, Thompson JR, Fuchs M, Perry KL. 2012. Complete genome sequence of a new circular DNA virus from grapevine. J Virol. 86:7715.
- Kuai YZ. 2010. Research progresses on mulberry virus and viral disease. Acta Sericol Sin. 36:818–825.
- Loconsole G, Saldarelli P, Doddapaneni H, Savino V, Martelli GP, Saponari M. 2012. Identification of a single-stranded DNA virus associated with citrus chlorotic dwarf disease, a new member in the family Geminiviridae. Virology. 432:162–172.
- Lu QY, Wu ZJ, Xia ZS, Xie LH. 2015. Complete genome sequence of a novel monopartite geminivirus identified in mulberry (Morus alba L.). Arch Virol. 160:2135–2138.
- Ma Y, Navarro B, Zhang Z, Lu M, Zhou X, Chi S, Di Serio F, Li S. 2015. Identification and molecular characterization of a novel monopartite geminivirus associated with mulberry mosaic dwarf disease. J Gen Virol. 96:2421–2434.
- Mansoor S, Khan SH, Bashir A, Saeed M, Zafar Y, Malik KA, Briddon RW, Stanley J, Markham PG. 1999. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology. 259:190–199.
- Mar TB, Mendes IR, Lau D, Fiallo-Olivé E, Navas-Castillo J, Alves MS, Zerbini FM. 2017. Interaction between the New World begomovirus Euphorbia yellow mosaic virus and its associated alphasatellite: effects on infection and transmission by the whitefly Bemisia tabaci. J Gen Virol. 98:1552–1562.
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1:1–5.
- Mubin M, Briddon RW, Mansoor S. 2009. Complete nucleotide sequence of Chili leaf curl virus and its associated satellites naturally infecting potato in Pakistan. Arch Virol. 154:365–368.
- Muhire BM, Varsani A, Martin DP. 2014. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 9:e108277.
- Paprotka T, Metzler V, Jeske H. 2010. The first DNA 1-like alphasatellites in association with New World begomoviruses in natural infections. Virology. 404:148–157.
- Polston JE, Londoño MA, Capobianco H. 2014. The complete genome sequence of New World jatropha mosaic virus. Arch Virol. 159:3131–3136.
- Qurashi F, Sattar M, Iqbal Z, Haider M 2017. First report of Cherry tomato leaf curl virus and associated DNA satellites infesting an invasive weed in Pakistan. J Plant Pathol. 99: 263–268.
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. 2005. Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol. 43:361–394.
- Romay G, Chirinos D, Geraud-Pouey F, Desbiez C. 2010. Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch Virol. 155:1843–1847.
- Shery AVMJ. 2016. A new variant of cassava mosiac virus causes mulberry mosaic disease in India. Int J Plant Animal Environ Sci. 6:83–92.
- Tahir M, Amin I, Haider S, Mansoor S, Briddon RW. 2015. Ageratum enation virus – a begomovirus of weeds with the potential to infect crops. Viruses. 7:647–665.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tsuchizaki T, Hibino H, Saito Y. 1971. Mulberry ringspot virus isolated from mulberry showing ringspot symptom. Jap J Phytopathol. 37:266–271.
- Wyatt SD, Brown JK. 1996. Detection of subgroup III geminivirus isolates in leaf extracts by degenerate primers and polymerase chain reaction. Phytopathology. 86:1288–1293.
- Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A. 2017. ICTV virus taxonomy profile: Geminiviridae. J Gen Virol. 98:131–133.