Abstract
Orthotospoviruses have become one of the major threats to vegetable crops worldwide. Recently, severe outbreaks of Tomato spotted wilt virus (TSWV) and Tomato yellow ring virus (TYRV) have been observed in tomato production regions in Poland. In this study, a multiplex RT-PCR assay was developed and optimized for the simultaneous detection of these two viruses. Primers were designed to amplify 666 bp and 328 bp of the nucleocapsid (N) genes of TYRV and TSWV, respectively. The multiplex RT-PCR detection limit was 1 pg μL−1 of total RNA, and it was successfully applied for the detection of TSWV and TYRV in 21 samples of tomato, tobacco and weed plants collected in Poland. Four samples were found to be co-infected with both viruses, whereas infection solely with TSWV and TYRV was found in 9 and 8 samples, respectively. Overall, this study presents a rapid, specific and sensitive method for orthotospoviruses identification in plants that can be applied to mixed infections.
Résumé
Les orthotospovirus sont devenus la principale menace pour les cultures maraîchères, et ce, à l’échelle planétaire. Récemment, plusieurs éclosions d’infections causées par le virus de la tache bronzée de la tomate (TSWV) et le virus des anneaux jaunes de la tomate (TYRV) ont été observées en Pologne, dans les régions productrices de tomate. Dans cette étude, une PCR multiplexe en temps réel a été développée et optimisée pour détecter simultanément ces deux virus. Des amorces ont été conçues pour amplifier 666 pb et 328 pb des gènes de la capside nucléique (N) de TYRV et de TSWV, respectivement. La limite de détection de cette PCR était de 1 pg μL−1 de l’ARN total, et elle a été utilisée avec succès pour détecter TSWV et TYRV dans 21 échantillons de tomate, de tabac et d’adventices collectés en Pologne. Quatre échantillons se sont avérés co-infectés par les deux virus, tandis que TSWV et TYRV ont infecté neuf et huit échantillons, respectivement. En résumé, cette étude présente une méthode rapide, précise et sensible qui permet d’identifier les orthotospovirus chez les plantes, et ce, dans les cas d’infections mixtes.
Introduction
Orthotospoviruses, belonging to the Tospoviridae family, are a serious threat to economically important plants worldwide (Pappu et al., Citation2009). The genus, named after the type member Tomato spotted wilt virus (TSWV), contains 11 approved and 18 tentative species (Chen & Jan, 2015; Adams et al., Citation2017). Orthotospoviruses are transmitted naturally in a persistent propagative manner by several thrips species (Plyusnin et al., Citation2011) and have a particular virion morphology with quasi-spherical particles measuring 80–120 nm in diameter. Each virus particle is bound by an envelope consisting of a host-derived membrane, which makes the orthotospoviruses unique among plant viruses (Timmerman-Vaughan et al., Citation2013). The virions contain a single copy of three single-stranded RNA molecules, denoted in relation to their size: large (L RNA), medium (M RNA) and small (S RNA). The L RNA contains an RNA-dependent RNA polymerase (RdRp) in a negative-sense orientation. In contrast, each of the M and S RNAs consists of two genes – one in the positive and the other in the negative-sense orientation. The M RNA encodes the NSm protein involved in cell-to-cell movement and the glycoprotein precursor, while the S RNA encodes the gene-silencing suppressor NSs and nucleocapsid (N) protein.
The widely occurring TSWV is able to infect more than 1090 plant species belonging to 85 families (Parrella et al., Citation2003). The virus causes a wide range of symptoms which depend on the host genotype, co-infections with other viruses, and environmental factors such as temperature (Pappu et al., Citation2009; Kulshrestha et al., Citation2013). In Poland, the presence of TSWV was reported in the early 1950s on tobacco plants (Żandarski & Kamińska, Citation1997). Since then, virus infection has been recorded in many crops, weeds and ornamental plants (Laskowska, Citation2008). The host range and geographic distribution of the established orthotospovirus species continue to increase (Pappu et al., Citation2009), and new species belonging to the Orthotospovirus genus have been proposed in recent years. One of them is the Tomato yellow ring virus (TYRV) identified for the first time in tomato plants in Iran (Hassani-Mehraban et al., Citation2005) and later on detected on subsequent agricultural and ornamental plants, such as potato, soya, chrysanthemum, gazania, cineraria, anemone and alstroemeria (Hassani-Mehraban et al., Citation2007; Rasoulpour & Izadpanah, Citation2007; Golnaraghi et al., Citation2008; Beikzadeh et al., Citation2012; Mortazavi et al., Citation2013). In 2015, TYRV was found for the first time on tomato in Poland (Zarzyńska-Nowak et al., Citation2016). The co-infection with TSWV was observed in 22% of the analysed samples. It is impossible to differentiate TSWV and TYRV based on the symptoms on infected plants; however, increased symptom severity was observed in cases of mixed infection (authors, unpublished observations). The development of molecular diagnostics offers the possibility for multiplex assay design by targeting several pathogens in a single reaction and discriminating between relatively closely related pathogens even within the same species.
Due to the fact that no chemical methods are available for curative treatment of plant viruses, it is important to prevent virus occurrence and create new plant protection strategies. Therefore, it is necessary to develop an effective and rapid detection method, especially for plant protection services trying to confine further spread of associated diseases. Several diagnostic techniques, such as enzyme-linked immunosorbent assay (ELISA), reverse transcription polymerase chain reaction (RT-PCR), quantitative RT-PCR, microarray, Luminex xTAG and reverse transcription loop-mediated isothermal amplification (RT-LAMP) have been developed for orthotospovirus detection (Bald-Blume et al., Citation2017; Fukuta et al., Citation2004; Balukiewicz et al., Citation2005; Liu et al., Citation2017; Huang et al., Citation2018). Conventionally, ELISA has been routinely used for large-scale screening of orthotospovirus infection in various types of plants (Charoenvilaisiri et al., Citation2014). However, these techniques are able to detect only individual orthotospovirus species in each reaction and are time-consuming and laborious, especially when large-scale diagnoses are performed. These limitations can be overcome by employing a multiplex RT-PCR (mRT-PCR) that can simultaneously amplify more than one target sequence of different viruses by several primer pairs in a single reaction, thereby offering a significant time and cost-saving advantage (Tuo et al., Citation2014). An mRT-PCR assay has been successfully applied for the detection of the following orthotospoviruses: TSWV, Impatiens necrotic spot virus (INSV), Chrysanthemum stem necrosis virus (CSNV), Iris yellow spot virus (IYSV), Capsicum chlorosis virus (CaCV) (Kuwabara et al., Citation2010) as well as TSWV, Watermelon silver mottle virus (WSMoV), Melon yellow spot virus (MYSV), WSMoV, INSV and IYSV (Uga & Tsuda, Citation2005). The goal of the present study was to develop and evaluate an mRT-PCR assay for the simultaneous detection and differentiation of TYRV and TSWV.
Materials and methods
Virus source
Four total RNA samples from tomato (Solanum lycopersicum) plants previously confirmed to be co-infected with TSWV and TYRV (Zarzyńska-Nowak et al., Citation2016) were used as templates for the optimization of uniplex RT-PCRs and mRT-PCR. RNA extracted from healthy tomato plants was used as negative controls. Total RNAs were isolated from 100 mg of infected plant leaves using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. The concentration of each RNA sample was measured in an ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and adjusted to 500 ng µL−1. Only the RNA samples with an A260/A280 ratio of 1.9–2.1 and an A260/A230 ratio greater than 2.0 were further used as a template in mRT-PCR reactions.
Primer design
The specific primer pairs were designed based on highly conserved regions of the full-length sequence alignment of nucleocapsid (N) protein gene of TSWV and TYRV isolates retrieved from the GenBank database (TSWV accession nos KR186203, KU949024.1, KU949025.1, KY569403.1, KX579057.1, LC167301.1, KU297996.1, KP330471.1, KP330470.1, KP330472.1, KM096536.1, HQ406903.1, HQ406942.1, HQ406948.1, HQ406956.1, KP008134.1, KP008131.1, KP008129.1, KF146701.1, KF146702.1, FJ234455.1); (TYRV accession nos KR139953-64, KX611798.1, KX611799.1, KX611800.1, JF836812.1, JF836810.1, FJ970490.1, DQ788693.1, DQ788694.1, DQ810195.1). Multiple sequence alignments were performed using ClustalW implemented in BioEdit Sequence Alignment Editor (Hall, Citation1999). Two different primer pairs were designed using OligoAnalyzer (www.uku.fi/~kuulasma/OligoSoftware) (). Subsequently, the specificity of each primer was evaluated using Primer-BLAST (Ye et al., Citation2012).
Table 1. Primers used in the multiplex detection of Tomato spotted wilt virus (TSWV) and Tomato yellow ring virus (TYRV).
The uniplex RT-PCR
The optimization of RT-PCR conditions is important for the development of a robust assay. In order to determine the optimum annealing temperature (Ta), the identical reaction condition with a fixed primer concentration was tested across a range of temperatures (Tuo et al., Citation2014). The reactions were performed using designed primer pairs and a Transcriptor One-Step RT-PCR Kit (Roche, Mannheim, Germany) in a T Professional thermocycler with a temperature gradient block (Biometra, Göttingen, Germany). The reactions were carried out in a 50 μL mixture containing 10 μL of 5× RT-PCR Reaction Buffer, 1 μL of Transcriptor Enzyme Mix, 2 μL of specific primer pairs (0.2 μM) (), 1 μL of extracted RNA and 34 μL of sterile water. Reverse transcription was performed at 50°C for 30 min. The initial denaturation was performed at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 10 s, annealing at 40 -55°C for 30 s, elongation at 68°C for 45 s, and a final cycle of 68°C for 7 min. In a negative control reaction, RNA isolated from healthy plants was used as a template. RT-PCR products were verified by electrophoresis on 1.5% agarose gel. In order to confirm the specificity of the obtained products, each amplified viral target was purified from gel, ligated into the pCR®4-TOPO vector (Thermo Fisher Scientific) and transformed into Escherichia coli TOP10 competent cells (Thermo Fisher Scientific). In the next step, plasmid DNAs were purified using NucleoSpin®Plasmid (NoLid) (Macherey-Nagel, Düren, Germany) and verified by EcoRI (Thermo Fisher Scientific) restriction. Plasmid DNAs were sequenced by an external company (Genomed S.A.,Warsaw, Poland). The obtained sequences were compared with others deposited in GenBank using BLASTn.
Optimization of mRT-PCR conditions
Among the two primer sets tested, the most promising results were obtained with TS1 and TY2. The mRT-PCR reactions were carried out using a different combination of primer concentrations. The mRT-PCR was performed using a Transcriptor One-Step RT-PCR Kit and a T Professional thermocycler with gradient options. Each reaction consisted of 10 μL of 5× Transcriptor Reverse Transcriptase Reaction Buffer, 1 μL of Transcriptor Enzyme Mix, 2 μL of 0.2 μM, 0.1 μM or 0.05 μM primers, 1 μL of extracted RNA and 30 μL of sterile water in a final volume of 50 μL. Reverse transcription was performed at 50°C for 30 min. The initial denaturation was performed at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 40 -55°C for 30 s, and elongation at 68°C for 30 s or 1 min. A final extension at 68°C for 5 min was also performed. The amplified RT-PCR products were subsequently separated by electrophoresis using 1.5% agarose gels.
The sensitivity of the uniplex and mRT-PCR assays
The sensitivity of the RT-PCR and mRT-PCR assays was determined using 10-fold dilutions of the extracted total RNA of plants co-infected with TSWV and TYRV. The RNA concentration was measured using an ND-1000 spectrophotometer, adjusted to 100 ng μL−1 and serially diluted from 100 ng μL−1 to 10 ag μL−1. RNA was then used as a template in RT-PCR and mRT-PCR reactions.
Detection of TSWV and TYRV in the field and greenhouse samples
Eighteen S. lycopersicum samples analysed in 2013–2014 at Plant Disease Clinic IPP-NRI and three samples (two from Nicotiana tabacum L. cv. Virginia and one from a weed plant – Melandrium album) provided by the Institute of Soil Science and Plant Cultivation, State Research Institute were simultaneously used to detect TSWV and TYRV by uniplex and mRT-PCR. RNA isolated from healthy tomato plants were used as the negative controls. The RNA extraction protocol was as described above.
Results
Specificity of the primer pairs
In uniplex RT-PCR, the primer combination TS1/TY2 gave clear and specific bands of target products compared with the other primer combination. The TS1/TY1 and TS2/TY2 primers combination gave non-specific products whereas no amplicons were obtained using the TS2/TY1 primer pair. TS1/TY2 primers performed well at temperatures ranging from 40°C to 50°C, and the products of the expected size of 666 bp and 328 bp were amplified for TYRV and TSWV, respectively. No products were obtained from healthy tomato plants. The obtained nucleotide sequences of TYRV and TSWV RT-PCR products confirmed the specificity of each primer pair. TS1 and TY2 primers were selected for further optimization in mRT-PCR.
Optimization of mRT-PCR conditions
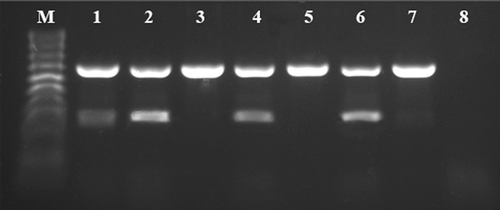
The gradient mRT-PCR reactions were performed across a range of temperatures, and the TS1/TY2 primers performed well between temperatures of 40°C and 50°C. Taking into account the amplification efficiency and specificity for the targeted viruses, the optimal annealing temperature was determined to be 45°C. For the combinations of different primer concentrations, the amplification efficiencies of TYRV and TSWV were evaluated (). A balanced amplification, with similar fluorescence intensity for the bands corresponding to the expected templates, was achieved when the primer concentrations were 0.1 μM for TYRV and 0.2 μM for TSWV (, line 2). In addition, there were no differences in band intensity between different RT-PCR elongation times. Ultimately, the optimized mRT-PCR cycle protocol was: reverse transcription at 50°C for 30 min, initial denaturation of 94°C for 7 min, 35 cycles of denaturation at 94°C for 45 s, annealing at 45°C for 30 s, elongation at 68°C for 45s and a final cycle of 68°C for 7 min. The 0.1 μM (TYRV) and 0.2 μM (TSWV) primers concentration were chosen as the optimal mRT-PCR reaction conditions.
Fig. 1 Optimization of the TY2/TS1 primer pairs concentration used for mRT-PCR. The assay was performed using total RNA isolated from a Solanum lycopersicum plants previously confirmed to be co-infected with Tomato yellow ring virus (TYRV) and Tomato spotted wilt virus (TSWV). PCR fragments of 666 bp and 328 bp are specific for TYRV and TSWV, respectively. M – HyperLadder™ 100 bp (Bioline). Lanes 1–7 indicate the concentration combinations of primer pairs specific for TYRV and TSWV, respectively: (1) 0.2:0.2 μM, (2) 0.1:0.2 μM, (3) 0.2:0.1 μM, (4) 0.05:0.1 μM, (5) 0.1:0.05 μM, (6) 0.05:0.2 μM, (7) 0.2:0.05 μM. Line 8: negative control.
Sensitivities of the uniplex and mRT-PCR assays
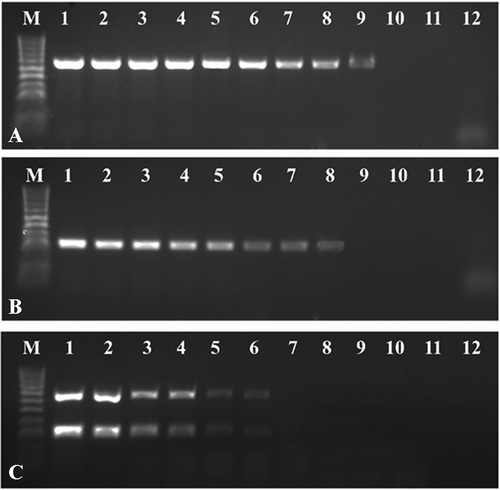
Sensitivities of uniplex RT-PCR and mRT-PCR for the detection of two orthotospoviruses were determined by using 10-fold serial dilutions of total RNA. The uniplex RT-PCR was able to detect an amount as low as 1 fg μL−1 and 10 fg μL−1 of TYRV and TSWV, respectively (,b), whereas the detection limit of the mRT-PCR was 1 pg μL−1 (). These results indicated that the multiplex RT-PCR was 100- or 1000-fold less sensitive than the uniplex RT-PCR but was still able to detect two viruses occurring in plant tissues in very low concentrations.
Fig. 2 Comparison of the sensitivities of uniplex and mRT-PCR assays for the detection of Tomato yellow ring virus (TYRV) and Tomato spotted wilt virus (TSWV). The uniplex and mRT-PCR assays were performed using total RNA isolated from a Solanum lycopersicum plants previously confirmed to be co-infected with both viruses and TY2/TS1 primer pairs. Fragments of 666 bp and 328 bp of TYRV and TSWV were amplified from 10-fold dilutions of total RNA starting at 100 ng μL−1. The figure depicting the electrophoretic separation of (a) RT-PCR TYRV products, (b) RT-PCR TSWV products and (c) mRT-PCR products. M – HyperLadder™ 100bp (Bioline). Lanes 1–11 – 10-fold dilutions of total RNAs, 12 – negative control.
Practical application of the assay for greenhouse and field samples
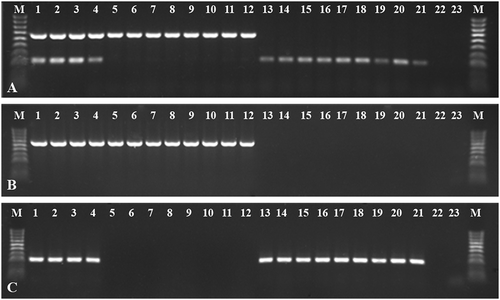
The optimized mRT-PCR method was used in practical application by detecting TYRV and TSWV in 21 plant samples. TYRV and TSWV mixed infection was confirmed in 4 samples from tomato, whereas infection solely with TYRV and TSWV was found in 8 tomato and 9 (6 tomato and 2 tobacco and 1 weed) plants, respectively (). All of these samples were further confirmed using uniplex RT-PCR, and the data showed that the results were in agreement with the mRT-PCR (,c), indicating that the mRT-PCR assay developed in this study is a rapid and validated method.
Fig. 3 Electrophoretic separations of mRT-PCR and uniplex RT-PCR products of 21 plant samples delivered to Institute of Plant Protection-NRI, Plant Disease Clinic (lanes 1–18 tomato plants, lanes 19–20 tobacco plants, lane 21 weed plant). (a) mRT-PCR detecting simultaneously both viruses, (b) and (c) uniplex RT-PCR Tomato yellow ring virus (TYRV) and Tomato spotted wilt virus (TSWV), respectively. M – HyperLadder™ 100 bp (Bioline). Lanes 1–21 tested samples, 22–23 negative controls.
Discussion
Orthotospovirus diagnosis plays an important role in crop improvement and disease management, especially when no effective curative treatment methods are available (EFSA, 2012). Orthotospoviruses are one of the most important groups of plant viruses responsible for economically important crop diseases in the Mediterranean (Turina et al., Citation2012). Recently, several outbreaks of TSWV in tomato and tobacco production regions have been reported in Poland. The occurrence of TSWV was also reported in wild plants, which can serve as a reservoir of the virus. Moreover, in 2016, the presence of TYRV was confirmed for the first time in Poland and Europe (Zarzyńska-Nowak et al., Citation2016). Both viruses cause yield and quality losses in greenhouse tomato and increased symptom severity was observed in plants co-infected with TSWV and TYRV. Taking into account the occurrence of orthotospoviruses in Poland, the designing of specific diagnostic tools and development of efficient and durable strategies for disease management is needed. Simultaneous detection of viruses from the Orthotospovirus genus was previously reported (Uga & Tsuda, Citation2005; Kuwabara et al., Citation2010; Charoenvilaisiri et al., Citation2014; Liu et al., Citation2017). Nevertheless, a technique for the detection of TSWV and TYRV, which can frequently be found together in mixed infections, was not available. Here, we developed a novel multiplex RT-PCR method for the simultaneous detection and differentiation of these two orthotospoviruses.
In mRT-PCR, the most important factors affecting the efficiency and specificity of the assay are yield and quality of extracted total RNA, and appropriative primer pairs (Uga & Tsuda, Citation2005; Tuo et al., Citation2014). We successfully designed virus-specific primers based on the nucleocapsid (N) protein gene sequence. The N protein is one of the most divergent orthotospoviral proteins, which has made it a major target for the taxonomic classification of orthotospoviruses (Chen et al., Citation2013). The sequences of this coding region were used to design specific primers in previously published diagnostic methods (Roberts et al., Citation2000; Okuda & Hanada, Citation2001; Kuwabara et al., Citation2010; Charoenvilaisiri et al., Citation2014; Liu et al., Citation2017).
In our study, we confirmed that the efficiency of the reaction was strongly dependent on the final concentrations of the primers. In optimized reaction conditions, the mRT-PCR developed was capable of detecting both viruses in as little as 1 pg µL−1 of total RNA. The comparison between uniplex and mRT-PCR assays revealed that mRT-PCR was 100- or 1000-fold less sensitive than the uniplex RT-PCR. Similar findings were reported in other studies (Uga & Tsuda, Citation2005; Panno et al., Citation2012; Tuo et al., Citation2014; Yao et al., Citation2014), which attribute it to some factor affecting mRT-PCR efficiency, such as primer competition for polymerase and templates (Nie & Singh, Citation2002). A problem in mRT-PCR is the unbalanced amplification of certain viruses due to the presence of multiple targets in one reaction and different primer compatibility to their targets, which may result in competition for enzymes and nucleotides in the reactions (Tuo et al., Citation2014). Nevertheless, the optimized mRT-PCR assay was 10–100 times more sensitive than other techniques described previously for TSWV detection, such as mRT-PCRs (TSWV detection limit of 20 pg μL−1) (Uga & Tsuda, Citation2005) or microarrays (TSWV detection limit of 100 pg μL−1) (Liu et al., Citation2017).
In conclusion, the mRT-PCR assay developed here provides a rapid and sensitive method for the simultaneous detection of TYRV and TSWV. To our knowledge, this is the first report of a multiplex one-step RT-PCR detection system for these two orthotospoviruses. The assay can serve as a suitable screening tool for the presence of TSWV and TYRV by phytosanitary protection services.
References
- Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, et al. 2017. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch Virol. 162:2505–2538.
- Bald-Blume N, Bergervoet JHV, Maiss E. 2017. Development of a molecular assay for the general detection of tospoviruses and the distinction between tospoviral species. Arch Virol. 162:1519–1528.
- Balukiewicz A, Kryczyński S, Golnik K. 2005. Biological differentiation and molecular characterization of Tomato spotted wilt virus (TSWV) isolates from chrysanthemum plants. Phytopathol Pol. 37:45–57.
- Beikzadeh N, Bayat H, Jafarpour B, Rohani H, Peters D, Hassani-Mehraban A. 2012. Infection of Alstroemeria plants with Tomato yellow ring virus in Iran. J Pathol. 160:45–47.
- Charoenvilaisiri S, Seepiban C, Bhunchoth A, Warin N, Luxananil P, Gajanandana O. 2014. Development of a multiplex RT-PCR-ELISA to identify four distinct species of tospovirus. J Virol Meth. 202:54–63.
- Chen T-C, Jan F-J. 2015. Tomato spotted wilt. In: Tennant P, Fermin G, editors. Virus diseases of tropical and subtropical crops. Wallingford (UK): CAB International; p. 167–176.
- Chen T-C, Li J-T, Fan Y-S, Yeh Y-C, Yeh S-D, Kormelink R. 2013. Molecular characterization of the full-length L an M RNAs of Tomato yellow ring virus, a member of genus Tospovirus. Virus Genes. 46:487–495.
- EFSA. European Food Safety Authority, Panel on Plant Health (PLH). 2012. Scientific opinion on the pest categorization of the tospoviruses. EFSA J. 10:3029.
- Fukuta S, Ohishi K, Yoshida K, Mizukami Y, Ishida A, Kanbe M. 2004. Development of immunocapture reverse transcription loop-mediated isothermal amplification for the detection of tomato spotted wilt virus from chrysanthemum. J Virol Meth. 121:49–55.
- Golnaraghi AR, Pourrahim R, Ahoonmanesh A, Zamani-Zadeh HR, Farzagfar S. 2008. Detection and characterization of a distinct isolate of Tomato yellow fruit ring virus from potato. Plant Dis. 92:1280–1287.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Sci. 41:95–98.
- Hassani-Mehraban A, Saaijer J, Peters D, Goldbach R, Kormelink R. 2005. A new tomato-infecting Tospovirus from Iran. Phytopathology. 95:852–858.
- Hassani-Mehraban A, Saaijer J, Peters D, Goldbach R, Kormelink R. 2007. Molecular and biologival comparison of two Tomato yellow ring virus (TYRV) isolate: challenging the Tospovirus species concept. Arch Virol. 152:85–96.
- Huang K-S, Li S-L, Sun J-H, Wang Y-C, Jan F-J, Chen T-C. 2018. Development of a generic method for inspection of tospoviruses. Eur J Plant Pathol. 150:457–469.
- Kulshrestha S, Sharma A, Seth CA. 2013. Molecular biology of Tomato spotted wilt virus: an update. J Appl Hort. 15:7180.
- Kuwabara K, Yokoi N, Ohki T, Tsuda S. 2010. Improved multiplex reverse transcription-polymerase chain reaction to detect and identify five tospovirus species simultaneously. J Gen Plant Pathol. 76:273–277.
- Laskowska D. 2008. Characterization of Tomato spotted wilt disease on tomato and vector role in transmission. Stud Rep Inst Soil Sci Plant Cultivation. 13:43–50.
- Liu L-Y, Ye H-Y, Chen T-H, Chen T-C. 2017. Development of a microarray for simultaneous detection and differentiation of different tospoviruses that are serologically related to Tomato spotted wilt virus. Virol J. 14:1.
- Mortazavi N, Aleosfoor M, Minaei K. 2013. Transmission of cineraria isolate of tomato yellow ring virus by Franliniella occidentalis and Thrips tabaci (Thysanoptera, Thripodae). Linzer Biol Beitr. 45:2011–2018.
- Nie X, Singh RP. 2002. A new approach for the simultaneous differentiation of biological and geographical strains of Potato virus Y by uniplex and multiplex RT-PCR. J Virol Meth. 104:41–54.
- Okuda M, Hanada K. 2001. RT-PCR for detecting five distinct Tospovirus species using degenerate primers and dsRNA template. J Virol Meth. 96:149–156.
- Panno S, Davino S, Rubio L, Rangel E, Davino M, García-Hernández J, Olmos A. 2012. Simultaneous detection of the seven main tomato-infecting RNA viruses by two multiplex reverse transcription polymerase chain reactions. J Virol Meth. 186:152–156.
- Pappu HR, Jones RA, Jain RK. 2009. Global status of tospovirus epidemics in diverse cropping systems: successes gained and challenges that lie ahead. Virus Res. 141:219–236.
- Parrella G, Gognalons P, Gebre-Selassie K, Vovlas C, Marchoux G. 2003. An update of the host range of Tomato spotted wilt virus. J Plant Pathol. 85:227–264.
- Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist A. 2011. Family bunyaviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, et al., editors. Ninth report of the international committee on taxonomy of viruses. Amsterdam (Netherlands): Elsevier Academic Press; p. 725–741.
- Rasoulpour R, Izadpanah K. 2007. Characterisation of cineraria strain of Tomato yellow ring virus from Iran. Australas Plant Path. 36:286–294.
- Roberts AA, Dietzgen RG, Heelan LA, Maclean DJ. 2000. Real-time RT-PCR fluorescent detection of tomato spotted wilt virus. J Gen Meth. 88:18.
- Timmerman-Vaughan GM, Lister R, Cooper R, Tang J. 2013. Phylogenetic analysis of New Zealand tomato spotted wilt virus isolated suggests likely incursion history scenarios and mechanisms for population evolution. Arch Virol. 159:993–1003.
- Tuo D, Shen W, Yang Y, Yan P, Li X, Zhou P. 2014. Development and validation of a multiplex reverse transcription PCR assay for simultaneous detection of three papaya viruses. Viruses. 6:3893–3906.
- Turina M, Tavella L, Ciuffo M. 2012. Tospoviruses in the mediterranean area. Adv Virus Res. 84:403–437.
- Uga H, Tsuda S. 2005. A one-step reverse transcription-polymerase chain reaction system for the simultaneous detection and identification of multiple tospovirus infections. Phytopathology. 95:166–171.
- Yao B, Wang G, Ma X, Liu W, Tang H, Zhu H, Hong N. 2014. Simultaneous detection and differentiation of three viruses in pear plants by a multiplex RT-PCR. J Virol Meth. 196:113–119.
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13:134.
- Żandarski J, Kamińska M. 1997. The problems of Tomato spotted wilt tospovirus (TSWV) in glasshouse and field crops in Poland-the occurrence, detection methods and control. Prog Plant Prot. 37:374–377.
- Zarzyńska-Nowak A, Rymelska N, Borodynko N, Hasiów-Jaroszewska B. 2016. The occurrence of Tomato yellow ring virus on tomato in Poland. Plant Dis. 100:234.
