Abstract
Narcissus (Narcissus spp.) is an important commercial crop and millions of narcissus bulbs are traded across the world annually. Three major viruses, namely, Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV) and Narcissus mosaic virus (NMV), have substantial impacts on global crop yields of narcissus. A multiplex RT-PCR assay was developed and used to simultaneously detect these three viruses. Three pairs of virus-specific primers were designed according to the conserved regions of the coat protein (CP) genes, generating expected fragments of 668 bp, 554 bp and 372 bp for NYSV, NLV and NMV, respectively. Specificity and compatibility of primer pairs were tested using uniplex and multiplex RT-PCR. Primer concentration, annealing temperature and cycle number were optimized to enable efficient and balanced amplification of the three viruses. The optimized multiplex RT-PCR was as sensitive as uniplex RT-PCR. Field samples were collected to validate the method, and all the viruses were successfully detected, thereby confirming the effectiveness and reliability of the developed multiplex RT-PCR. This method could be used for large-scale surveys of these three viruses in narcissus and thus might play a role in diagnosis and prevention of these pathogens.
Résumé
Le narcisse (Narcissus spp.) est une importante culture commerciale et des millions de bulbes sont échangés annuellement dans le monde. Trois principaux virus, à savoir le virus de la bigarrure jaune du narcisse (VBJN), le virus latent du narcisse (VLN) et le virus de la mosaïque du narcisse (VMN) ont des répercussions substantielles sur les rendements mondiaux des cultures de narcisse. Un biotest basé sur la PCR multiplexe a été développé et utilisé pour détecter simultanément ces trois virus. Trois paires d’amorces spécifiques des virus ont été conçues en fonction des zones conservées des gènes de la protéine de l’enveloppe, engendrant des fragments attendus de 668 bp, de 554 bp et de 372 bp pour VBJN, VLN et VMN, respectivement. La spécificité et la compatibilité des paires d’amorces ont été testées par PCR uniplexe et multiplexe. La concentration des amorces, la température d’hybridation et le nombre de cycles ont été optimisés pour permettre une amplification efficace et équilibrée des trois virus. La PCR multiplexe optimisée s’est avérée aussi sensible que la PCR uniplexe. Des échantillons ont été collectés au champ pour valider la méthode et tous les virus ont été détectés avec succès, confirmant ainsi l’efficacité et la fiabilité de la nouvelle PCR multiplexe. Cette méthode pourrait être utilisée pour des études de grande envergure sur ces trois virus chez le narcisse et pourrait ainsi contribuer au diagnostic et à la prévention de ces agents pathogènes.
Introduction
Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV) and Narcissus mosaic virus (NMV) are three major viruses identified in narcissus. NYSV, a member of the Potyvirus genus in Potyviridae family, is found worldwide and is generally more prevalent than the others. This virus can be transmitted to healthy plants by aphids and causes a series of symptoms inducing streaks on leaves, distortion of leaves, retarded growth of the diseased plant and declines in yields. It is also involved in the phenomenon of breaking flowers (Brunt, Citation1971). NLV belongs to Macluravirus genus that infects narcissus and occasionally some bulbous plants including irises, gladiolus and nerines (Brunt, Citation1977). It is common in Poland and has been reported from some commercial cultivation fields in New Zealand and Japan (Clark & Guy, Citation2000; Sochacki, Citation2011; Hori et al., Citation2013). Plants infected with NLV alone usually show inconspicuous symptoms, making it impossible to relate specific leaf symptoms to virus infection. The filamentous NMV is a member of the genus Potexvirus first found in British cultivars, later in Otago Province New Zealand, in Poland and recently in Australia in iris (Brunt, Citation1966; Clark & Guy, Citation2000; Sochacki, Citation2011; Wylie et al., Citation2014). Typical symptoms for NMV infection include mosaic and distortion on the leaves. Recently, research showed the presence of NMV was associated with the colour break in reverse bicolour daffodils (Narcissus pseudonarcissus) (Hunter et al., Citation2011). In several surveys, samples infected with NLV were coincidentally infected with NYSV and/or NMV, and it was suspected that NLV accelerated the effects of other viruses (Brunt, Citation1971, Citation1977; Clark & Guy, Citation2000).
Enzyme-linked immunosorbent assay (ELISA) and reverse transcription polymerase chain reaction (RT-PCR) have been employed to detect these viruses from narcissus (Clark & Guy, Citation2000; Sochacki, Citation2011; Hori et al., Citation2013; Berniak et al., Citation2013). ELISA, as a classic serological method for virus detection, possesses a number of advantages such as simplicity and the ability to test a large number of samples. However, the low sensitivity and limited numbers of commercially available antibodies for the virus, to some extent, restricts its application. Unlike ELISA, RT-PCR is sensitive and specific (Berniak et al., Citation2013). On the other hand, RT-PCR is inefficient in dealing with large number samples or mixed-infection, which is common in narcissus plants. Multiplex RT-PCR is an assay that simultaneously amplifies several RNA targets in a tube containing corresponding primer pairs. Since multiplex RT-PCR is time-saving and cost-efficient, it has been widely used to detect and/or differentiate viruses infecting dahlias (Asano et al., Citation2015), lily (Kwon et al., Citation2013), orchid (Ali et al., Citation2014), canna (Chauhan et al., Citation2015) and some other plants. (Wei et al., Citation2009; Li et al., Citation2012; Panno et al., Citation2012; Noorani et al., Citation2013; Yu et al., Citation2013; Zhang et al., Citation2013; Tuo et al., Citation2014; Nam et al., Citation2015; Hao et al., Citation2016; Yang et al., Citation2017). However, it is relatively difficult to detect more than three plant viruses in a multiplex RT-PCR assay, probably due to the technical difficulties of a reaction involving so many compatible primers (López et al., Citation2009).
Previous studies have demonstrated the mixed infection of NYSV, NLV and/or NMV in narcissus using conventional methods (Brunt, Citation1971, Citation1977; Clark & Guy, Citation2000). Recently, Hori et al. (Citation2013) developed a multiplex RT-PCR assay to investigate the distribution of NYSV and NLV in production fields in Fukui Prefecture Japan. In our previous study, an efficient TaqMan-based multiplex qRT-PCR was developed to detect NYSV and NMV in narcissus (Jin et al., Citation2017). Both of these studies found mixed-infection of NYSV/NLV and NYSV/NMV. However, to date, no molecular detection method has been reported for detecting three narcissus viruses (e.g. NYSV, NLV and NMV) simultaneously. Herein, we developed a multiplex RT-PCR assay for simultaneous detection of NYSV, NLV and NMV with the aim of providing a simple, rapid and reliable method for large-scale survey of three viruses in port quarantine and agricultural production. To the best of our knowledge, this is the first report on the simultaneous detection of NYSV, NLV and NMV using multiplex RT-PCR.
Materials and methods
Plant materials
Narcissus leaf tissue samples infected with NYSV, NLV and NMV were identified using conventional RT-PCR (Shen et al., Citation2012a, Citation2012b, Citation2014) and stored in the lab at −80°C. The samples were used to establish the multiplex RT-PCR method. For the field samples originating from different geographic regions, bulbs were purchased from cultivation fields in Zhangzhou and Pingtan, China and the Netherlands and planted in a test field in Fuzhou, Fujian province. Leaf tissues from these samples were gathered and subjected to the optimized multiplex RT-PCR for virus diagnosis.
Design of primers
Complete or partial genomic sequences of NYSV isolates (accession numbers: KU516386, KP893337, KM066972, JQ686724, EU430294, AM158908, AJ311372), NLV isolates (accession numbers: KP893334, JX270762, JN662384, JX270761, JN615214, JN615212, FJ024083, JN662386, JX270765) and NMV isolates (accession numbers: KP893336, JQ911733, KF752593, AY225449, D13747) were retrieved from GenBank and aligned with Clustal X to obtain the conserved regions in coat protein (CP) genes. The software Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA) was used to design the virus-specific primers, check the annealing temperatures and evaluate the probability of intermolecular or intramolecular interactions. Details of the analysed primers are provided in .
Table 1. Primers employed in the multiplex RT-PCR assay for detection of Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV) and Narcissus mosaic virus (NMV).
RNA extraction and cDNA synthesis
Approximately 100 mg of virus-infected or virus-free narcissus leaf tissue per sample were manually homogenized with a mortar and pestle in the presence of liquid nitrogen. Plant RNA solutions were prepared using a TransZol Up Plus RNA kit (TransGen Biotech, Beijing, China) following the manufacturer’s protocol and stored at −80°C. The quantity and quality of the extracted RNA was analysed by measuring absorptions at 260 nm and 280 nm using Thermo NanoDrop 2000c (Thermo Scientific, Massachusetts, USA). The RNA was transcribed into cDNA using the following protocol: incubation of an 11-μL reaction mixture containing 7 μL DEPC-treated water, 1 μL hexamer random primer (Thermo Scientific) and 3 μL RNA (~0.5 μg) at 70°C to allow the primers to bind to the RNA. Next, the addition of another 9-μL mixture containing 5 μL 5×buffer (Promega, Wisconsin, USA), 2 μL dNTP mix (Promega), 1 μL RNAsin Plus RNase inhibitor (Promega), and 1 μL M-MLV reverse transcriptase (Promega). The mixture was incubated at 42°C for 60 min and subsequently at 70°C for 10 min. The cDNAs were stored at −20°C until use.
Uniplex RT-PCR
For uniplex RT-PCR, a 25 μL mixture containing 2 μL cDNA, 12.5 μL 2× GoTaq Green Master mix (Promega), 0.25 μL forward and backward primers (10 μM each) and 10 μL distilled water was prepared in microtubes. The tubes were placed in a cycler (Veriti® 96-Well Thermal Cycler, ABI, USA) for amplification of the target genes with pre-denaturation at 94°C for 3 min, followed by 30 cycles of 30 s denaturation at 94°C, 30 s annealing at 48–58°C and 45 s extension at 72°C and then a final extension at 72°C for 10 min. Products were separated by electrophoresis in a 1.2% agarose gel and visualized using gel imaging analysis system (G:BOX Chemi XRQ, Syngene, UK). To further confirm the specificity of the primers, amplicons were cloned and then sequenced. The sequences were subjected to BLAST search on GenBank database.
Multiplex RT-PCR
For the initial multiplex RT-PCR, individual viral RNAs of similar concentrations were artificially mixed and added into the cDNA mix. An aliquot of 2 μL cDNA was added to the reaction containing 12.5 μL GoTaq Green Master Mix, and 0.25 μL of each primer (10 μM). Various combinations of primer pairs were tested and distilled water was added to obtain a final volume of 25 μL. The amplification program was the same as that for uniplex RT-PCR (annealing temperature at 52°C). The amplicons were analysed using electrophoresis and an UV transilluminator.
To optimize the multiplex RT-PCR assay, primer concentration, annealing temperature and number of cycles were examined. According to the results of the initial multiplex RT-PCR, primer concentrations of NMV were set within 0.02−0.08 μM, NYSV from 0.3 to 0.5 μM, while NLV remained at 0.1 μM. The annealing temperature was determined at 48–58°C with increments of 2°C. Cycles of 25, 30, 35 and 40 were tested. For primer concentrations, the combination that produced a balanced amplification of all the target genes was considered the optimal. For the annealing temperature, the one with proper amplification efficiency was selected for further experiments, whereas for cycle number, the one that gave balanced amplification efficiency and elapsed time for the assay was determined to be optimal.
Cloning and sequencing of PCR products
The amplified product of multiplex RT-PCR was gel purified using a PCR purification kit (Tiangen, Beijing, China). The expected fragments were cloned into the pMD18-T simple vector, transformed into Escherichia coli JM109 and sequenced in both directions by Sangon Biotech Co., Ltd (Shanghai, China).
Sensitivities of uniplex and multiplex RT-PCR
To determine the detection limit of the established multiplex RT-PCR assay, total RNA extracted from narcissus plants singly infected with NYSV, NLV and NMV was mixed and diluted serially 10-fold (10°–10−6) in nuclease-free water. The diluted RNA solutions were converted into cDNA and subjected to uniplex and multiplex RT-PCR assay. The sensitivities of the assays were then compared.
Field sample detection using multiplex RT-PCR
A total of 148 samples with relevant symptoms or asymptomatic originating from Zhangzhou and Pingtan in China’s Fujian province or the Netherlands were tested using the optimized multiplex RT-PCR. Products were separated by electrophoresis and virus-positive samples were scored.
Results
Specificity and compatibility of the primers
Specificity of the primer pairs was separately tested by uniplex RT-PCR with individual viral cDNA using an annealing temperature of 48–58°C in increments of 2°C. All the targets were successfully amplified with expected bands at 668 bp for NYSV, 554 bp for NLV and 372 bp for NMV (data not shown). Annealing temperatures from 52–56°C consistently amplified all the targeted regions of the viruses; therefore, a temperature of 52°C was employed for further experiments (data not shown). No amplification of the negative control or cross-reaction was observed, suggesting the specificity of the primers to their targets. PCR products were sequenced and subjected to a BLASTn search on the GenBank database, and high homology to the published NYSV, NLV and NMV sequences were obtained. These findings further confirmed the conclusion that the primers were specific to their targets.
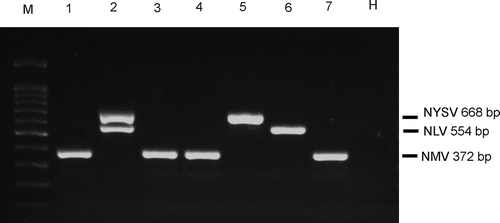
The initial multiplex RT-PCR was carried out using 2 μL cDNA and all possible combinations of the primer pairs in equal volumes. As shown in , NYSV, NLV and NMV were efficiently amplified using one pair of the virus-specific primers (, lanes 5, 6 and 7). In the tubes containing NMV and NYSV/NLV primers, amplification of NYSV or NLV was inhibited, indicated by the absence of the bands at 668 bp for NYSV and 554 bp for NLV on the gel after electrophoresis (, lanes 3 and 4). It is intriguing to note that in the tube contained NYSV and NLV primers both of the targets were successfully amplified ( lane 2), while in the sample with three sets of primers, only NMV was amplified ( lane 1). A possible explanation for these results could be the competitive inhibition of NMV primers against primers for NYSV and NLV.
Fig. 1 Multiplex RT-PCR detection of Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV) and Narcissus mosaic virus (NMV) with three sets of primer pairs. M, 100 bp DNA marker (Tiangen, Beijing, China); Lane 1, NYSV +NLV +NMV; lane 2, NYSV +NLV; lane 3, NYSV +NMV; lane 4, NLV +NMV; lane 5, NYSV only; lane 6, NLV only; lane 7, NMV only; H, healthy leaf.
Optimization of multiplex RT-PCR
To obtain an unbiased amplification of NYSV, NLV and NMV, primer concentrations were first tested. Since NMV was preferentially amplified, its primer concentration was gradually decreased to 0.02 μM, while the others remained at 0.1 μM. The results revealed that amplification of NYSV and NLV recovered as the primer concentration of NMV dropped to 0.032 μM and below. Furthermore, NLV and NMV had similar amplification efficiency when primer concentrations were 0.1 μM for NLV and 0.032 μM for NMV. We further tested the primer concentration of NYSV ranging from 0.2 μM to 0.5 μM. The balanced amplification was achieved when the primer concentrations were 0.3 μM for NYSV, 0.1 μM for NLV and 0.032 μM for NMV (data not shown). The annealing temperature from 48 to 58°C was tested in a gradient cycler. The results showed the temperature had limited influence on the amplification efficiencies of the viruses. Equal amplification of all the targets was obtained at 52°C. Higher temperature resulted in a slight decrease of the amplification efficiency for NMV and an increase of that for NYSV and NLV (data not shown). The number of cycles tested was 25, 30, 35 and 40. Amplification efficiency of NMV was stable throughout the cycles. However, NYSV and NLV were effectively amplified when the cycle number was increased to 30; more cycles resulted in more amplicons and a longer reaction time (data not shown). Therefore, 30 cycling number was chosen for the following experiments. In summary, the optimal conditions were primer concentrations at 0.3 μM for NYSV, 0.1 μM for NLV, 0.032 μM for NMV and 30 cycles with denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 45 s.
Sequence analysis of PCR products
The multiplex RT-PCR products were sequenced and compared with BLASTn, confirming that the positive amplicon sequences shared more than 98% identity with the reported sequences of NYSV, NLV and NMV in GenBank.
Sensitivity of uniplex and multiplex RT-PCR
Sensitivity of uniplex and the optimized multiplex RT-PCR was determined using serial 10-fold dilution of the mixed total RNA solution. In uniplex RT-PCR, the detection limits of NYSV, NLV and NMV were 10−4, 10−3 and 10−3, respectively (, and ). In the multiplex RT-PCR, 10−4 dilution of NYSV could be successfully detected and the detection limits for NLV and NMV were 10−2 and 10−3, respectively (). The results indicated that the sensitivity of the optimized multiplex RT-PCR was similar to that of uniplex RT-PCR.
Fig. 2 The sensitivities of uniplex and multiplex RT-PCR assays for Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV) and Narcissus mosaic virus (NMV). The PCR assays were conducted using serial dilution of the mixed total RNA, which were converted into cDNA to serve as the templates. M, 100 bp DNA marker (Tiangen, Beijing, China); a, uniplex RT-PCR amplification of NYSV; b, uniplex RT-PCR amplification of NLV; c, uniplex RT-PCR amplification of NMV; d, multiplex RT-PCR amplification of NYSV, NLV and NMV.
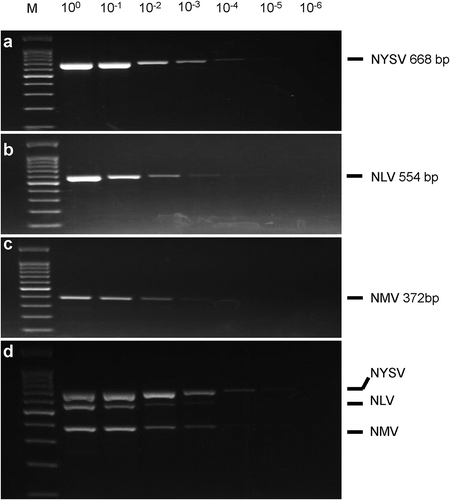
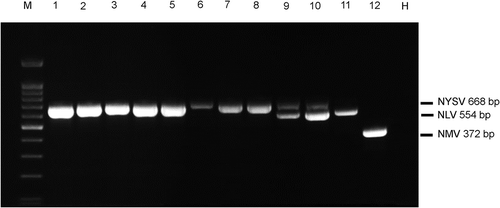
Fig. 3 Detection of 12 samples with conspicuous symptoms on the leaves of cultivars collected from Zhangzhou, Pintan and the Netherlands using the optimized multiplex RT-PCR of three pairs of virus-specific primers for Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV) and Narcissus mosaic virus (NMV). M, 100 bp DNA marker (Tiangen, Beijing, China); lanes 1–4, samples from Zhangzhou; lanes 5–8, samples from Pintan; lanes 9–12, samples from the Netherlands; H, healthy sample.
Evaluation of multiplex RT-PCR
To validate the optimized multiplex RT-PCR, 12 samples with virus-specific symptoms on the leaves were collected from different geographic regions and subjected to the multiplex PCR system. The result showed the presence of all three viruses in the samples from the Netherlands but only NYSV in the samples from Zhangzhou and Pingtan ().
An additional 136 samples with or without symptoms were tested to further evaluate the assay. As shown in , 123 out of 148 samples (83.1%) were infected with at least one virus, among which 115 samples (77.7%) were infected with a single virus. Specifically, 104 samples (70.3%) were infected with NYSV only, 9 samples (6.1%) with NLV only and 2 samples (1.4%) with NMV only. Mixed infection was not common in these samples since only 8 (5.4%) were found mix-infected with NYSV/NLV. Moreover, mixed infections with NYSV/NMV, NLV/NMV and NYSV/NLV/NMV were not observed. NYSV was predominant in all of the cultivation fields in Fujian Province, China and the Netherlands, while NLV and NMV were only identified in the samples from the Netherlands. The detection results of field samples were further verified using uniplex RT-PCR, and the results were identical with those of multiplex RT-PCR, suggesting that the established multiplex RT-PCR can effectively be used for routine detection of the three viruses.
Table 2. Analysis of 148 narcissus samples with or without symptoms on the leaves for Narcissus yellow stripe virus (NYSV), Narcissus latent virus (NLV)and Narcissus mosaic virus (NMV) using the optimized multiplex RT-PCR.
Discussion
Narcissus is a popular cut flower that is grown all over the world. It is an important commercial crop for some developing countries and a host for up to 24 viruses, which influence the quality and quantity of this plant (Brunt, Citation1971; Berniak et al., Citation2013). NYSV, NLV and NMV are three major viruses in narcissus that tend to form complexes in cultivars from several production fields (Brunt, Citation1971, Citation1977; Clark & Guy, Citation2000). Reliable and effective methods to simultaneously detect these viruses are limited. In the present paper, we developed a multiplex RT-PCR assay using three sets of specifically designed primers targeting to NYSV, NLV and NMV. Primer concentration, annealing temperature and cycle number were tested to optimize the assay. All of the targets were successfully detected under optimal conditions and the sensitivity was comparable to uniplex RT-PCR. The results of field sample testing further validated this method.
In multiplex RT-PCR, special attention should be paid to primer design since chances for primer interaction increases when two or more primer pairs are included in the reaction mixture. Apart from avoiding forming secondary structures, ideal primers for multiplex RT-PCR are supposed to have similar annealing temperatures to enable high amplification efficiency and to produce amplicons that differ by approximately 100 bp in size to ensure separable bands on the gel (Elnifro et al., Citation2000; Wei et al., Citation2008). In this study, three sets of primers producing amplicons of 668 bp, 554 bp and 372 bp were employed (). Gradient uniplex RT-PCR using annealing temperatures at 48, 50, 52, 54, 56 and 58°C was conducted to test the primers and only the target bands were observed, suggesting specificity of the primers. However, in the initial multiplex RT-PCR, amplification of NYSV and/or NLV was significantly inhibited by NMV (). Changing annealing temperature slightly altered the amplification efficiency of the primers and 52°C was considered optimal for all the targets (data not shown).
Unbalanced amplification is common in multiplex RT-PCR (Elnifro et al., Citation2000) and preferential amplification of NMV against NYSV and/or NLV occurred when equimolar primer pairs were included in the reaction (). To address this issue, we adjusted the concentrations of primer pairs by decreasing that of NMV to 0.02 μM and kept those of NYSV and NLV unchanged (0.1 μM). When primer concentration of NMV was at 0.032 μM, similar amplification efficiency for NLV and NMV were achieved while signal for NYSV was weak, which was indicated by the dim fluorescence intensity of the 668 bp band. Primer concentration of NYSV was gradually increased to 0.5 μM, and balanced amplification of the three viruses was achieved when the primer concentrations were 0.3 μM for NYSV, 0.1 μM for NLV and 0.032 μM for NMV. The detection limit of the multiplex RT-PCR was similar with that of uniplex RT-PCR. Previous researchers have claimed an ~10- to 100-fold decline in sensitivity compared to uniplex RT-PCR and this reduction has been attributed to competition between primer pairs in the systems (Yu et al., Citation2013; Tuo et al., Citation2014; Hao et al., Citation2016; Yang et al., Citation2017).
It is important to note that several groups included internal controls in the multiplex PCR systems to guarantee the quality of RNA template (Wei et al., Citation2009; Kwon et al., Citation2013; Noorani et al., Citation2013). However, it has some disadvantages such as competition between amplification of internal control and target genes, and might not work for every species of plant (Zhang et al., Citation2013). In this study, no internal control was added to the reaction because the quality was immediately analysed after RNA extraction, and only those with high quality were kept. Second, all the viruses (NYSV, NLV, NMV) were specifically amplified with relatively high amplification efficiency by uniplex RT-PCR (data not shown), suggesting the RNA was of good quality. Finally, competitive inhibition is common during multiple amplifications of target genes and as Zhang et al. (Citation2013) have described, the primers of internal controls might act as competitors for reaction resources. This competition could result in reduced sensitivity or even an inability to diagnose multi-virus infected samples.
A total of 148 field samples from Zhangzhou, Pingtan and the Netherlands were used to validate the assay. Among the virus-positive samples, most of them were infected with NYSV, which further confirmed the general observation that occurrence of the virus is higher than other narcissus viruses worldwide (Brunt, Citation1971; Clark & Guy, Citation2000). NLV and NMV only existed in the samples from the Netherlands. Detection rate of NMV was low as only two Netherlands-based samples were NMV-positive. Mixed-infection of NYSV and NLV was observed in samples from the Netherlands. The result was in accordance with a previously published survey’s conclusion that NLV tends to form complexes with NYSV (Brunt, Citation1971). In conclusion, a rapid, specific and sensitive multiplex RT-PCR assay for simultaneous detection of NYSV, NLV and NMV in narcissus has been established. This assay has the potential to be used as an efficient tool for large-scale surveys and certification programmes.
Additional information
Funding
References
- Ali RN, Dann AL, Cross PA, Wilson CR. 2014. Multiplex RT-PCR detection of three common viruses infecting orchids. Arch Virol. 159:3095–3099.
- Asano S, Matsushita Y, Hirayama Y, Naka T. 2015. Simultaneous detection of Tomato spotted wilt virus, Dahlia mosaic virus and Chrysanthemum stunt viroid by multiplex RT‐PCR in dahlias and their distribution in Japanese dahlias. Lett Appl Microbiol. 61:113–120.
- Berniak H, Komorowska B, Sochacki D. 2013. Detection of Narcissus latent virus isolates using one-step RT-PCR assay. J Hort Res. 21:11–14.
- Brunt AA. 1966. Narcissus mosaic virus. Ann Appl Biol. 58:13–23.
- Brunt AA. 1971. Virus diseases of Narcissus. In: Daffodil and tulip year book. Roy Hort Soc; p. 18–37.
- Brunt AA. 1977. Some hosts and properties of Narcissus latent virus, a carlavirus commonly infecting narcissus and bulbous iris. Ann Appl Biol. 87:355–364.
- Chauhan RP, Wijayasekara D, Webb MA, Verchot J. 2015. A reliable and rapid multiplex RT-PCR assay for detection of two potyviruses and a pararetrovirus infecting canna plants. Plant Dis. 99:1695–1703.
- Clark V, Guy P. 2000. Five viruses in Narcissus spp. from New Zealand. Australas Plant Path. 29:227–229.
- Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 13:559–570.
- Hao L, Xie JP, Chen SY, Wang SJ, Gong ZQ, Ling KS, Guo LY, Fan ZF, Zhou T. 2016. A multiple RT-PCR assay for simultaneous detection and differentiation of latent viruses and apscarviroids in apple trees. J Virol Methods. 234:16–21.
- Hori E, Sakamoto H, Ohki S, Imura R, Goshima Y. 2013. Establishment of a virus assay system by RT-PCR for Narcissus tazetta var. chinensis and the distribution of virus infection in culture fields. Acta Hortic. 1002:237–241.
- Hunter DA, Fletcher JD, Davies KM, Zhang H. 2011. Colour break in reverse bicolour daffodils is associated with the presence of Narcissus mosaic virus. Virol J. 8:412.
- Jin J, Shen JG, Cai W, Xie GH, Liao FR, Gao FL, Ma JF, Chen XH, Wu ZJ. 2017. Narcissus yellow stripe virus and Narcissus mosaic virus detection in Narcissus via multiplex TaqMan‐based reverse transcription‐PCR assay. J Appl Microbiol. 122:1299–1309.
- Kwon JY, Ryu KH, Choi SH. 2013. Reverse transcription polymerase chain reaction-based system for simultaneous detection of multiple lily-infecting viruses. Plant Pathol J. 29:338–343.
- Li F, Zuo RJ, Abad J, Xu DL, Bao GL, Li RH. 2012. Simultaneous detection and differentiation of four closely related sweet potato potyviruses by a multiplex one-step RT-PCR. J Virol Methods. 186:161–166.
- López MM, Llop P, Olmos A, Cambra M, Bertolini E. 2009. Are molecular tools solving the challenges posed by detection of plant pathogenic bacteria and viruses? Curr Issues Mol Biol. 11:13–46.
- Nam M, Lee YH, Park CY, Lee MA, Bae YS, Lim S, Lee JH, Moon JS, Lee SH. 2015. Development of multiplex RT-PCR for simultaneous detection of garlic viruses and the incidence of garlic viral disease in garlic genetic resources. Plant Pathol J. 31:90–96.
- Noorani MS, Awasthi P, Sharma MP, Ram R, Zaidi AA, Hallan V. 2013. Simultaneous detection and identification of four cherry viruses by two step multiplex RT-PCR with an internal control of plant nad5 mRNA. J Virol Methods. 193:103–107.
- Panno S, Davino S, Rubio L, Rangel E, Davino M, García-Hernández J, Olmos A. 2012. Simultaneous detection of the seven main tomato-infecting RNA viruses by two multiplex reverse transcription polymerase chain reactions. J Virol Methods. 186:152–156.
- Shen JG, Gao FL, Cai W, Yu WT, Yu Y, Li M, Yan C. 2012a. Establishment and application of RT-PCR for detection of Narcissus mosaic virus. Chin Agric Sci Bull. 28:206–210.
- Shen JG, Gao FL, Lin SQ, Li M, Yu WT, Yu Y, Cai W. 2014. Detection of Narcissus yellow stripe virus by one-step RT-PCR. Biotechnology. 24:44–48.
- Shen JG, Gao FL, Yu Y, Cai W, Li M, Yu WT, Liao FR. 2012b. Detection of Narcissus latent virus by RT-PCR. Chin J Trop Crops. 33:1808–1811.
- Sochacki D. 2011. The use of ELISA in the micropropagation of virus-free narcissus. Acta Hortic. 886:253–258.
- Tuo D, Shen WT, Yang Y, Yan P, Li XY, Zhou P. 2014. Development and validation of a multiplex reverse transcription PCR assay for simultaneous detection of three papaya viruses. Viruses. 6:3893–3906.
- Wei T, Lu GJ, Clover G. 2008. Novel approaches to mitigate primer interaction and eliminate inhibitors in multiplex PCR demonstrated using an assay for detection of three strawberry viruses. J Virol Methods. 151:132–139.
- Wei T, Lu GJ, Clover G. 2009. A multiplex RT‐PCR for the detection of Potato yellow vein virus, Tobacco rattle virus and Tomato infectious chlorosis virus in potato with a plant internal amplification control. Plant Pathol. 58:203–209.
- Wylie SJ, Li H, Liu J, Jones MG. 2014. First report of Narcissus mosaic virus from Australia and from Iris. Australas Plant Dis Notes. 9:134.
- Yang X, Lv K, Wang MQ, Zhou GH. 2017. Investigation of viruses infecting rice in southern China using a multiplex RT-PCR assay. Crop Prot. 91:8–12.
- Yu Y, Zhao Z, Jiang D, Wu Z, Li S. 2013. A one‐step multiplex RT‐PCR assay for simultaneous detection of four viruses that infect peach. Lett Appl Microbiol. 57:350–355.
- Zhang JQ, Wang R, Song JZ, Luo ZP, Yang J, Lin FC. 2013. One‐step multiplex RT-PCR for simultaneous detection of four viruses in tobacco. J Phytopathol. 161:92–97.
