Abstract
Surveys of pulse crops across the Canadian prairies were undertaken between 2014 and 2017 to assess the distribution and severity of root rot of field pea and lentil, to identify the most important causal agents, and to compare the efficiency of culturing techniques with multiplex PCR. Root rot was present in every pea and lentil field surveyed in all three provinces. When assessed based on province and soil zone, moderate to severe symptoms (severity > 3 on a 1–7 scale) occurred in 25–99% of pea fields, but only 8–34% of lentil fields. Root rot severity on both crops was higher in growing seasons with high precipitation and was lower in drier years, but only lentil showed any difference associated with soil zone, with lower severity in the brown soil zone. This study demonstrated that Aphanomyces euteiches occurs across the Prairie region, where it caused moderate to severe injury on many field pea crops. Fusarium avenaceum was the most common pathogen identified in both culturing and PCR analyses, but other Fusarium spp., together with Pythium spp. and Rhizoctonia solani, were also components of the complex. Use of PCR for pathogen identification routinely resulted in identification of many pathogens from an infected root, whereas culturing and isolation resulted in only one or two pathogens per root. PCR was extremely important for assessing the impact of A. euteiches, which was almost never identified using culturing approaches. This observation explains why A. euteiches was not initially recognized as an important pathogen of pulses in the region.
Résumé
De 2014 à 2017, des études sur les légumineuses ont été entreprises dans les Prairies canadiennes pour évaluer la distribution et la gravité du pourridié chez le pois potager et la lentille, et ce, afin d’en identifier les principaux agents causaux et de comparer l’efficacité des techniques de culture à celle de la PCR multiplex. Le pourridié a été détecté dans tous les champs de pois et de lentilles étudiés dans les trois provinces. Lorsqu’ils ont été évalués en fonction de la province et de la zone pédologique, des symptômes légers à graves (gravité > 3 sur une échelle de 1 à 7) ont été décelés dans 25 à 99% des champs de pois, mais seulement de 8 à 34% dans ceux de lentilles. La gravité du pourridié à l’égard des deux cultures était plus intense durant les saisons de croissance affichant de forts taux de précipitations, et moindre durant les saisons plus sèches. Par contre, seules les lentilles ont affiché une différence associée à la zone pédologique, présentant une plus faible gravité dans la zone de sol brun. Cette étude a démontré qu’Aphanomyces euteiches colonise toutes les Prairies où il provoque des lésions mineures à graves dans de nombreuses cultures de pois potagers. Fusarium avenaceum était l’agent pathogène le plus fréquemment identifié par culture et par PCR, mais d’autres espèces de Fusarium, de concert avec Pythium spp. et Rhizoctonia solani, appartenaient au complexe. Le recours à l’analyse par PCR pour détecter les agents pathogènes a permis d’identifier couramment de nombreux agents sur une racine infectée, tandis que la culture et l’isolement ont permis d’en identifier seulement un ou deux par racine. La PCR a été des plus importantes pour évaluer la portée d’A. euteiches qui n’était presque jamais détecté par les analyses basées sur la culture. Cette constatation explique pourquoi A. euteiches n’avait pas été initialement reconnu en tant qu’agent pathogène majeur chez les légumineuses dans la région.
Introduction
Canada is the world’s largest exporter of field pea (Pisum sativum L.) and lentil (Lens culinaris L.), producing ~3.9 M metric tonnes (MT) of pea and 2.3 MT of lentil each year, mostly for export. Production is concentrated in the Prairie region, with large acreages in Saskatchewan and Alberta, and a smaller area of pea production in Manitoba (Statistics Canada, Citation2017). Root rot of pulses, especially field pea, has been increasing on the Canadian prairies in recent years (Gossen et al., Citation2016). Yield losses caused by root rot are difficult to quantify, but complete yield loss has been observed in heavily infested pea fields in Alberta and Saskatchewan (Banniza et al., Citation2013; Chatterton et al., Citation2015b).
Disease surveys on pulse crops have been made frequently in Saskatchewan and Manitoba, but have been based primarily on rating of plants in the field, and generally did not determine the main pathogen(s) associated with root rot. Surveys that include a diagnostic component have not been conducted across the major pulse growing regions of Alberta since at least 2000, and have never been done on lentil. Surveys were conducted in the black soil zone near Edmonton, Alberta in late July 2007 (Feng et al., Citation2010), but the pea production area in Saskatchewan and Alberta has expanded and shifted, with a high proportion of the acreage now occurring in the brown and dark brown soil zones of the southern prairies.
Recent studies of root rot pathogens of pulses in North Dakota indicated that pathogen populations were changing over time (Chittem et al., Citation2015), probably associated with changes in agronomic practices and crop diversity. It is likely that the predominant root pathogens of pea on the Canadian prairies have also changed since they were last studied, given the shifts that have occurred in the main production areas and changes in cropping practices over the last 20 years.
Root rot of pea is caused by a pathogen complex, with Fusarium solani and F. avenaceum implicated as primary causal organisms in recent studies using culture-based isolation techniques (Feng et al., Citation2010; Chittem et al., Citation2015; Esmaeili Taheri et al., Citation2017a). However, culture-based methods for isolation of plant pathogenic organisms have inherent biases based on choice of sample processing steps, tissue sterilization procedure, and culture media (O’Brien et al., Citation2005). It is also a time-consuming process when dealing with a pathogen complex and with a large number of samples. Aphanomyces euteiches, which is an important oomycete pathogen of pea globally (Pfender & Hagedorn, Citation1983; Gaulin et al., Citation2007), was recently reported from Alberta and Saskatchewan and was only detected using PCR-based identification from diseased pea roots (Armstrong-Cho et al., Citation2014; Chatterton et al., Citation2015a). It was also detected at relatively high abundance in soil samples collected from pea fields in Alberta, Saskatchewan and Manitoba (Esmaeili Taheri et al., Citation2017b).
The objectives of this study were to determine the incidence and severity of pea root rot on the Canadian prairies and to identify the predominant Fusarium spp. associated with root rot using a multiplex PCR. After the first year, the objectives of the study were expanded to assess the distribution of A. euteiches on field pea and lentil, which are the most important pulse crops on the Canadian prairies.
Materials and methods
Weather data
Precipitation data from May–August each year were obtained from Environment Canada for AAFC Research Centres in soil zones surveyed in each province. Although this approach uses a single site to represent a large geographic region, it allows examination of broad differences in major weather patterns among years and regions.
Surveys
To assess the prevalence, incidence and severity of root rot on pulse crops, fields were surveyed at flowering for above- and below-ground symptoms of root rot. Surveys were conducted in commercial pea fields in Alberta in 2014. In 2015, surveys were expanded to pea crops in Saskatchewan and lentil crops in Alberta. In 2016 and 2017, surveys were expanded again to include pea crops in Alberta, Saskatchewan and Manitoba, lentil crops in Alberta and Saskatchewan, and dry bean, faba bean and alfalfa crops in Alberta. Surveys were predominantly performed during early-mid flowering (early-mid July), but there was some variation in this timing across years and provinces.
In 2014, there was a major outbreak of root rot in Alberta; ~50% of the fields included in the survey were assessed because a producer reported crop collapse in their field, and the remainder of fields were selected arbitrarily. In 2015–2017, crops were selected arbitrarily, but focused on locations of field pea production within a county or crop district. Crops were evaluated at 10 sites per field along a U-shaped pattern, with a minimum of 30 m between sites. The GPS and/or legal land location of each field was collected at the field entry point. At each site, plants within a 1-m-long section of row were rated for above-ground symptoms of root rot using a 1–5 scale, where 1 = healthy plants; 2 = slight yellowing of lower leaves; 3 = yellowing of leaves up to 4th node; 4 = yellowing of more than 50% of the plant, and 5 = complete yellowing or all plants dead (Infantino et al., Citation2006).
To assess root rot severity, roots from 5–10 plants were dug up at each of the 10 sampling sites per field, bagged per site and stored at ~ 4° C for up to a week prior to processing. Samples from Manitoba, Saskatchewan and central Alberta were shipped on ice in cooler packs to the Lethbridge Research and Development Centre for assessment. Roots were washed under running tap water for 10 min, and then individual roots were assigned a visual rating for root rot severity on the 1–7 scale described in (Bilgi et al., Citation2008).
Table 1. Description of the visual rating scale (1–7) used to assess root rot incidence and severity in laboratory assessments (slightly modified from Bilgi et al., Citation2008).
Roots collected in early June in southern Alberta in 2016–2017 from fields with symptoms of severe root rot were used for isolation of A. euteiches. Lateral roots were examined for presence of oospores under a microscope, and roots with oospores were plated without surface sterilization onto a semi-selective medium for A. euteiches. The medium consisted of cornmeal agar amended with metalaxyl, benomyl and vancomycin (MBV) (Pfender et al., Citation1984). In addition, other roots from samples with a severity rating of 4–7 were processed for DNA extraction.
Pathogen detection using PCR
Roots were pooled from each site per field, the whole root cut into 0.5-cm-long pieces, and placed into 15-mL Falcon tubes. In instances where all of the sites within a field had very high and uniform root rot ratings, roots from 2–3 sites were pooled to produce three replicates per field, rather than test all 10 sites individually. Tubes were frozen in liquid N, and then stored at −80°C prior to freeze-drying (LabConco, Kansas City, MO) for 48 h. Aliquots of freeze-dried root tissue (30 mg) were transferred to 8-strip collection microtubes in a 96-well plate format (Qiagen, Carlsbad, CA). Samples were ground using a TissueLyzer II (Qiagen) and 3-mm tungsten-carbide beads (Qiagen). DNA was extracted using the BioSprint Plant DNA kit (Qiagen) according to manufacturer’s instructions, except that the amount of buffer RLT added to the ground tissue was increased from 300 to 500 µL. To ensure that this increased volume did not affect the quality of the DNA extracts, random samples (10 per plate) were checked for DNA quantity and quality using a Qubit 4 Fluorometer (ThermoFisher) according to the manufacturer’s instructions. DNA was used in a series of multiplex reactions to test for the presence of eight Fusarium species commonly isolated during initial screening. The multiplex reactions were performed using the Qiagen Multiplex PCR Kit according to manufacturer’s directions, with 2 μL of DNA, 0.2 mM of each primer, and inclusion of the optional Q-solution for a final 20 μL reaction. Reactions were run on an Eppendorf MasterCyclerS as follows: Fusarium/Aphanomyces reactions – Initial heat activation – 15 min at 95°C; 30 s at 94°C, 90 s at 60°C, 60 s at 72°C for 35 cycles; followed by a final extension for 10 min at 72°C; Pythium/Rhizoctonia reactions – Initial heat activation – 15 min at 95°C; 30 s at 94° C, 30 s at 60°C, 60 s at 72°C for 35 cycles; followed by a final extension for 10 min at 72°C.
To develop the multiplex assays, literature was reviewed for primers specific to each species of interest, ordered from IDT (Coralville, IA), and tested in singleplex reactions against all species of interest, as well as other common soil-borne fungi that had been isolated in culturing experiments (Esmaeili Taheri et al., Citation2017a). DNA for positive standards was extracted from fungal cultures using the DNeasy Plant Kit (Qiagen) according to manufacturer’s directions. Standard cultures were isolated previously from diseased pea roots and identification was confirmed using morphology and DNA sequencing (Esmaeili Taheri et al., Citation2017a). Once acceptable primers had been identified, primers targeting combinations of species were tested. There was cross-reactivity against F. acuminatum and F. tricinctum for all of the F. avenaceum primer pairs tested. Assessments that combined the reactions for both F. avenaceum and F. acuminatum generally differentiated these two species, but in some cases the identification was not clear and so was scored as mixed avenaceum/acuminatum. Initial combinations were based on expected amplicon size, and Tm of primers (data not shown). Multiplex combinations were as follows: (1) F. graminearum, F. oxysporum and A. euteiches + EF; (2) F. acuminatum, F. solani and F. equisiti; (3) F. culmorum, F. redolens and A. euteiches; and (4) F. avenaceum (2 primer pairs) + ITS ().
Table 2. Description of the primer sequences (with references) used to identify individual root rot pathogens.
ITS and EF were added to the multiplex assays to serve as universal positive controls for Fusarium spp. and general fungi, and served as an indicator of a positive PCR reaction in the event that no target species were amplified in a reaction. If the ITS or EF PCR failed and there was no amplification of any of the target species, the sample was not included in final summaries. A positive DNA standard from stock cultures of all species was included with every multiplex reaction run on each batch of root DNA samples to ensure there was no cross-reactivity between primer pairs. In 2016 and 2017, assessment of Pythium ultimum, P. irregulare and Rhizoctonia solani were added to the analysis.
PCR results were visualized on a 1.25% agarose gel using EzVision One loading dye (Amresco, Solon, OH) according to manufacturer’s instructions. Agarose gels were run at 90V for 70 min in 0.5× TBE solution, and visualized using a BioRad GelDoc System (BioRad Laboratories, Mississuaga, ON). If a DNA standard did not amplify as expected in a given reaction, the reaction was repeated. Bands were scored according to expected band size relative to the positive control size and intensity (strong, weak or negative), and were only scored as positive or negative if the positive and negative DNA standards also gave the appropriate response. PCR for A. euteiches was performed in duplicate for each sample. Samples that did not result in the same duplicate response were repeated a third time, and the consensus result (2/3) was taken as the correct result. All other reactions were not performed in duplicate, but there were always multiple samples tested per field.
Culturing versus multiplex PCR
A subset of 21 fields from the 2014 and 2015 surveys were chosen for in-depth sampling, to compare the efficiency of identification of common root rot pathogens using a standard culturing approach against multiplex PCR. Methods and results for culturing were described previously (Esmaeili Taheri et al., Citation2017a). Fields that showed a large variation in disease rating between sites (i.e. diseased patches) were chosen for the in-depth study, with at least one field assessed in each soil zone where field pea is grown in each province. The same samples of roots and stems used for culturing were processed and analysed using multiplex PCR, as described above. The only difference was that five root samples per site were analysed for each of the 10 collection sites per field using PCR, regardless of the root rot rating at that site. The frequency of detection of specific pathogens using each method was summarized and compared.
Statistical analysis
Fields surveyed were categorized according to soil zone (black, dark brown, brown) for analysis of root rot incidence and severity. Soil zones were assigned using the land location on the Alberta Agriculture and Forestry website, or by location in Saskatchewan. Any locations in the thin black or black-dark grey areas were considered as black soil zone locations for analysis. There were a small number of fields surveyed in the grey soil zone in Saskatchewan, the Peace River soil zone in Alberta, or a soil zone could not be assigned due to a missing location. These fields were excluded from statistical analysis.
Root rot severity > 3 was arbitrarily identified as a level that would potentially impact root function because the tap root was girdled and loss of root mass was observed above this rating level. The incidence of samples (sites) with mean severity > 3 was calculated to assess the frequency of moderate to severe root rot. Field prevalence was determined as the proportion of fields where at least one site had root rot.
PCR reactions were scored as 1 (positive) or 0 (negative), and used to calculate the proportion of fields and samples that were positive for each pathogen. It was not possible to test all of the root samples from all fields because of constraints imposed by project logistics and economics, so only samples with a root rating > 3 were included in the PCR assays. However, the total number of fields surveyed was used to calculate the proportion of fields that were positive for each pathogen. Under these constraints, the roots with a rating < 3 were, by definition, negative (even though they were in fact only less likely to be positive for any given pathogen than roots > 3). For calculating mean proportion of positive samples, only fields that were included in the DNA screening were included in the analysis. The effect of year, soil zone and year × soil zone were analysed using proc glimmix in SAS 9.4 (SAS Institute Inc.) for root rot severity, and root rot incidence > 3. These variables were considered as pseudo-continuous variables at the population level (McRoberts et al., Citation2003). Variances were unequal between provinces, and thus data for each province were analysed separately. Homogeneity of variance assumptions for year and soil were tested by plotting studentized residuals in SAS, and a heterogeneous error covariance structure applied to the model when necessary to fix unequal variances (Bowley, Citation2015). To satisfy assumptions of normality and random distribution, disease severity data were log-transformed and incidence data were arcsine square-root transformed using the link function. Means were separated using Tukey’s HSD and differences are significant at P ≤ 0.05 unless otherwise stated. Means and standard errors were back-transformed using the ilink term in the SAS glimmix model (Bowley, Citation2015).
The frequency of positive fields and samples of each organism for each province was treated as nominal data and thus mean and standard error was calculated using the tabulate function in JMP 12.0. The effect of year, soil and soil × year on A. euteiches frequency by field was analysed using the Generalized Linear Model platform in JMP 12.0 (SAS Institute Inc.) with a binomial distribution, logit function and maximum likelihood estimation method for each province. When an effect was significant, significant differences were determined using the contrast function for each individual parameter. The same analysis was performed for per cent fields positive for each Fusarium species, except, for simplicity, only the effect of year was considered in the analysis. The association between shoot rating and root rot severity rating by pea or lentil crop at each site was determined using the Fit Y-by-X platform in JMP 12.0, as each variable was considered to be pseudo-continuous.
To determine a crude distribution pattern of A. euteiches across sites within fields, number of samples (1 pooled root sample/site) tested within a given field were assigned to five categories: 0, 1–25%, 26–50%, 51–75% and 76–100% samples positive, and the proportion of fields within each category calculated by province (Alberta or Saskatchewan), year and crop (pea or lentil).
Results
Weather data
Analysis of general precipitation patterns across the Prairie Provinces from 2014 to 2017 revealed that the growing seasons of 2014 and 2016 were generally wetter than the long-term average (LTA) for each location, while 2015 and 2017 were much drier than the LTA (). This was particularly evident in the brown soil zone (Swift Current, SK and Lethbridge, AB), which had the largest deviations from the LTA. Precipitation generally occurred late in the growing season (August–September) in 2016, which resulted in a wet spring in some locations in 2017. These locations, however, received little to no rain in July–August.
Table 3. Total rainfall (mm) from May–August each year relative to the 30-year long-term average (LTA) at locations in the brown, dark brown and black soil zones on the Canadian prairies, 2014–2017.
Surveys
Occasionally symptoms on pea roots presented as classical Aphanomyces root rot symptoms (honey-brown discolouration of lateral roots, epicotyl pinching and cortical decay), Fusarium root rot (blackened tap root and reddening of the vascular bundle), but most often symptomatology was not diagnostic for one causal agent (,,). Symptoms on lentil were more difficult to diagnose and rate (). Above-ground symptom expression also varied greatly and above-ground symptoms were not always a good indicator of root rot severity. For example, early-season symptoms of yellowing and stunting of young shoots were common in 2016 and 2017, particularly in regions where there was adequate moisture in the spring (). In 2014, extensive collapse of entire pea and lentil fields were visible by early July, in regions that received above-average rainfall in June (). In contrast, symptoms in 2015, in regions that experienced a relatively dry growing season, appeared as yellowing patches following a rain event during or after flowering (). Correlation analysis of shoot ratings with root rot severity ratings at each site showed that there was a weak correlation for pea (r = 0.21, P < 0.001) but none for lentil (r = 0.013, P = 0.05).
Fig. 1. (Colour online) Root rot symptoms: (a) severe root decay and shoot death of pea caused by A. euteiches; (b) vascular reddening and blackening of tap root caused by Fusarium spp.; (c) honey-brown discolouration and epicotyl pinching caused by A. euteiches; (d) lentil roots showing, from left to right, healthy, moderate and severe root symptoms; (e) early season symptoms of Aphanomyces root rot on pea shoots causing yellowing and stunting in 2017; (f) yellowing patches developing after a mid-July rainfall in 2015; (g) pea field showing severe yield loss on one half, and patchy distribution along depressions and water tracks on the other half in 2014.

There was no effect of soil zone or zone × year on root rot incidence or severity for pea in Alberta and Saskatchewan. In Manitoba, pea production occurs only in the black soil zone, so the effect of soil type could not be tested. However, there were differences among years on some variables in Alberta and Saskatchewan, so means are presented for each year. Root rot incidence and severity on pea in Alberta was significantly higher in 2014 than in 2015 (P = 0.0220 and 0.0428, respectively), 2016 (P = 0.0001 and 0.0011, respectively) and 2017 (P = 0.0047 and 0.011), which were not significantly different from each other (P values ranged from 0.3452–1.000). In Saskatchewan in 2015, pea root rot incidence and severity was significantly higher than in 2016 and 2017 (P < 0.0001 for all comparisons), which were similar (P = 0.9977 and 0.6658, respectively) (). There were no differences in root rot incidence or severity between 2016 and 2017 in Manitoba (P = 0.1162 and 0.1149, respectively) (). Moderate to severe symptoms (severity > 3) were present in 25–50% of fields, except Saskatchewan in 2015, where fields were surveyed late and severity was high (, ). As such, mean severity ranged from 2.6–3.9, with Manitoba generally showing the highest severity and Alberta the lowest (), although these were not compared statistically.
Table 4. Number of fields surveyed, and mean root rot incidence (at damaging levels, DI > 3) and severity, of pea and lentil fields in Alberta, Saskatchewan and Manitoba, 2014–2017.
Fig. 2. (Colour online) Average root rot severity of each pea or lentil crops surveyed in Alberta (2014–2017), Saskatchewan (2015–2017) and Manitoba (2016–2017).
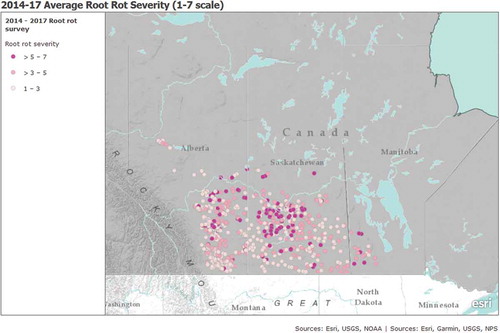
Root rot was also present in every lentil field surveyed (). For lentil, virtually all production in Alberta occurs in the brown soil zone, and there is little or no lentil production in other soil zones, so there was no possibility of an interaction with soil zone. In Alberta, there was no significant difference between root rot severity among years (P = 0.7812), but there was a difference in incidence among years. Incidence was significantly lower in 2017 than 2016 (P = 0.0223), but was similar to incidence in 2015 (P = 0.058), which was also similar to 2016 levels (P = 0.5294) (). In Saskatchewan, incidence and severity were significantly higher in 2016 than in 2017 (P < 0.0001; ). There was also an effect of soil zone on lentil in Saskatchewan for both incidence (P = 0.002) and severity (P < 0.0001). Root rot levels in the black (incidence = 0.31, severity = 2.9) and dark brown (0.18, 2.7) soil zones were similar (P = 0.5520 and 0.2813, respectively), but were both significantly higher compared with the brown soil zone (0.09, 2.2; P = 0.0002 and 0.0026 (incidence); P < 0.0001 (severity)) (data not shown).
Pathogen detection using PCR: Aphanomyces euteiches
Aphanomyces euteiches was identified across the Prairie Provinces wherever pea and lentil crops were grown, and as far north as the Peace River region in Alberta (). There was no soil zone or soil zone × year interaction for the frequency of pea fields positive for A. euteiches in Saskatchewan (P = 0.4123 and 0.4855, respectively). In Alberta, there was an effect of soil zone (P = 0.0098), but not years (P = 0.4792), or an interaction effect (P = 0.055), with 40–51% of pea fields positive for A. euteiches in every year (). The black and dark brown soil zones had a higher proportion of fields positive for A. euteiches (51% and 50%, respectively) than the brown soil zone (36%), and this difference was significant based on contrast analysis (P = 0.012 and 0.007, respectively). There were differences in the frequency of pea fields positive for A. euteiches among years in both Saskatchewan (P = 0.0052) and Manitoba (P = 0.024) (,e). In Saskatchewan, 69% of pea fields were positive for A. euteiches in 2015 and 65% in 2016, which was higher than in 2017 (40%) (P = 0.003 and 0.009). In Manitoba, the proportion of positive fields was higher in 2016 (73%) than in 2017 (45%) (P = 0.024).
Fig. 3. (Colour online) Aphanomyces euteiches incidence and per cent positive samples (sites) based on PCR assays from diseased root tissues, within pea and lentil crops in Alberta (2014–2017), Saskatchewan (2015–2017) and Manitoba (2016–2017).
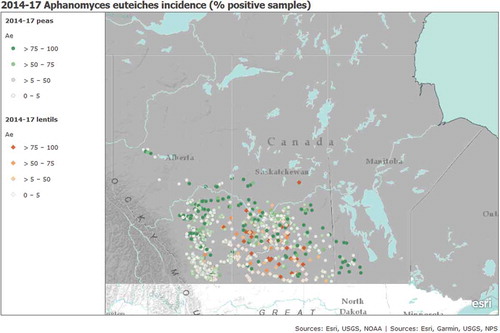
Fig. 4. Per cent fields and samples (sites) testing positive for Aphanomyces euteiches, based on PCR assays from diseased root tissues, and total precipitation over the growing season (May–August) in (a) Alberta pea; (b) Alberta lentil; (c) Saskatchewan pea; (d) Saskatchewan lentil; and (e) Manitoba pea crops in their respective years surveyed. Error bars represent standard error of the mean.
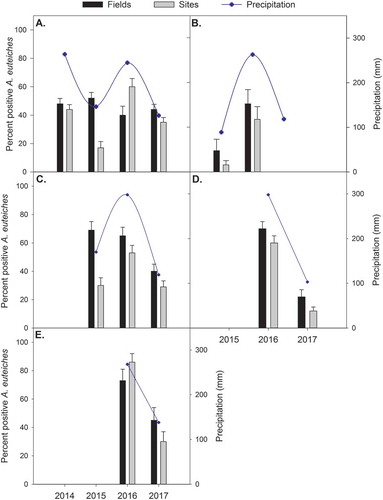
In Alberta, the frequency of positive samples was lowest in 2015 (18%), intermediate in 2014 (43%) and 2017 (36%), and highest in 2016 (61%) (). There were also differences in soil zone, with the black and dark brown zones having the highest frequency of positive samples (53% and 42%) compared with the brown soil zone (23%) (data not shown). In Saskatchewan, the frequency of positive samples in 2015 (26%) and 2017 (26%) were lower than in 2016 (57%) (), but levels were similar across soil zones (data not shown). In Manitoba, the proportion of positive samples was higher in 2016 (86%) than in 2017 (30%) ().
On lentil in Alberta, 15% of fields were positive for A. euteiches in 2015 and 48% in 2016 which were significantly different (P = 0.0163). There were no positive fields detected in 2017, and this was not significantly different from 2015 (P = 0.05) but was from 2016 (P = 0.0001) (). Only 5% of samples were positive in 2015, but this increased to 37% in 2016. On lentil in Saskatchewan, there was a significant effect of year (P < 0.0001), but not soil (P = 0.2710) or their interaction (P = 0.1817) on proportion of positive fields. The proportion of fields positive for A. euteiches was significantly higher in 2016 than in 2017 (70% vs. 22% of fields, P < 0.0001) ().
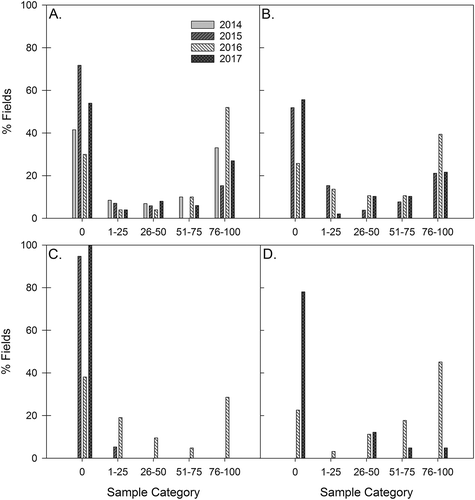
The distribution of positive and negative A. euteiches samples within a field showed a bimodal distribution, with the majority of samples within a given field testing either all negative or all positive (). For pea, in Alberta in 2014 and 2016, and Saskatchewan in 2016, 33–52% of fields had 76–100% of samples test positive for A. euteiches, while in 2015 and 2017, 15–25% of fields were in the 76–100% category for positive samples (,). For lentil in 2016 in Alberta 20% of fields were in the 1–25% category, 5% in 26–50%, and 30% in 76–100% (). There were very few positive lentil fields in 2015 and all (5%) were in the 1–25% category. For lentil in Saskatchewan in 2016, the largest proportion of fields (45%) was in the 76–100% category, followed by 51–75% (18%) and 26–50% (11%) (). In 2017, there were few fields that were positive for A. euteiches, but of those that tested positive, 12% of fields had 26–50% of samples positive for A. euteiches, followed by 5% in both the 51–75% and 76–100% categories.
Fig. 5. The proportion of (a) Alberta pea; (b) Saskatchewan pea; (c) Alberta lentil and (d) Saskatchewan lentil fields in each category, based on the % samples from each field that tested positive for Aphanomyces euteiches in PCR analyses, 2014–2017 (NB only includes fields with root rot severity ratings > 3).
Isolation from Alberta root samples was attempted in 2015, 2016 and 2017 with the aim of creating an A. euteiches culture collection. Despite a concentrated effort, only 3, 14 and 3 isolates were obtained in 2015, 2016 and, 2017, respectively. Soil baiting and isolations were not performed due to the number of fields that would have needed sampling and testing.
Pathogen detection using PCR: Fusarium, Pythium and Rhizoctonia
The overall results for F. avenaceum and F. acuminatum were combined, and are presented as F. avenaceum in . In pea in Alberta, there were differences among years for all Fusarium spp., except F. graminearum which was low in each year (). Generally, a higher proportion of fields were positive for Fusarium spp. (avenaceum, solani, redolens, oxysporumand culmorum) in 2015 and 2017 than in 2014 or 2016 (; Supplementary Table 1).
Fig. 6. Per cent fields testing positive for important Fusarium spp., based on PCR assays from diseased root tissues, in (a) Alberta pea; (b) Alberta lentil; (c) Saskatchewan pea; (d) Saskatchewan lentil; and (e) Manitoba pea crops in their respective years surveyed. Error bars represent standard error of the mean.
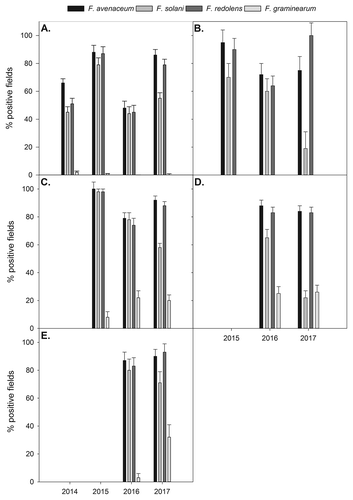
A similar trend was observed on pea in Saskatchewan. Fusarium avenaceum and F. redolens were the dominant species with 80–100% of positive fields (; Supplementary Table 1). There were lower levels of F. solani positive fields in 2017 (58%) compared with 2015 (98%) or 2016 (78%). The incidence of Fusarium graminearum and F. culmorum was higher in 2017 and 2016 compared with 2015, but proportions of positive fields were generally lower than for the other Fusarium spp. In Manitoba, there were no differences in the Fusarium spp. composition between the 2 years, except that F. graminearum had higher levels in 2017 than in 2016 (; Supplementary Table 1).
On lentil, F. avenaceum, F. redolens and F. oxysporum were the predominant root rot pathogens in Alberta and Saskatchewan, found in 72–100% of fields each year, except for F. redolens in Alberta in 2016 (,; Supplementary Table 2). Fusarium solani was also common (60–65% of fields), but was significantly lower in 2017 than 2015 or 2016 in both Alberta and Saskatchewan. Fusarium graminearum was not detected on lentil roots from Alberta, but was present in 25% of fields in Saskatchewan in both 2016 and 2017. Fusarium culmorum was present in 19–43% of fields, and frequency did not differ between years.
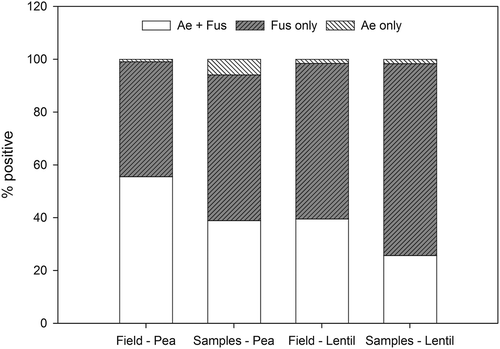
Of the fields and samples that tested positive for A. euteiches, F. avenaceum or F. solani, the proportion that tested positive for single or combinations of these pathogens were calculated (). The majority of pea samples (55%), and lentil fields (59%) and samples (73%), were positive for Fusarium only (F. avenaceum and/or F. solani), while 55% of the total pea fields were positive for A. euteiches and at least one of the two Fusarium species. Very few pea and lentil fields and samples were positive for A. euteiches in the absence of F. avenaceum or F. solani.
Fig. 7. Per cent of pea and lentil fields and samples (sites) that were positive for either A. euteiches and F. avenaceum and/or F. solani (Ae+ Fus); F. avenaceum and/or F. solani (Fus only); or A. euteiches (Ae only).
Pythium ultimum and P. irregulare were present in 8–43% of pea fields in Alberta, with frequency of detection higher in 2017 compared with 2016 (Supplementary Table 1). In Saskatchewan and Manitoba, 16–49% of fields were positive, with no significant differences between years. Similar results were also observed for lentil (Supplementary Table 2). Rhizoctonia solani was frequently detected in both pea and lentil fields in all three provinces (Supplementary Table 1, 2).
Culturing versus multiplex PCR
Results of isolations onto culture media from root samples from a subset of pea fields assessed in 2014 and 2015 (Esmaeili Taheri et al., Citation2017a) were compared with the results of the multiplex PCR assays to assess the strengths and weakness of each approach. For example, A. euteiches was not detected in cultures from any root sample, but was detected in 37% of samples in 2014 and 45% in 2015 (). The frequency of detection of Fusarium spp. using PCR was always higher than the frequency of isolation into culture for all species. The only exception was F. acuminatum, where the detection frequency was similar for both methods. Generally, one to two species were recovered from each root using isolation into culture, whereas PCR detected four to five species per sample. On the other hand, F. torulosum and F. tricinctum were recovered in culture but were not included in the PCR assays because primers specific to these species were not available. Pythium species were also commonly isolated in culture, so P. ultimum and P. irregulare were added to the PCR assays in 2016. Rhizoctonia solani was not frequently isolated in culture, but this species was also added to the PCR assays in 2016 since it can be a common soil-borne pathogen of pea (Hwang et al., Citation2007).
Table 5. Frequency of identification of specific root rot pathogens based on assessment using culturing versus multiplex PCR, based on comparison of results from a subset of surveyed fields. Five fields in each year were chosen that had visible healthy and diseased patches, and were within 100 km of research centres.
Other legume crops in Alberta
A small number of alfalfa, faba bean and dry bean fields were surveyed in Alberta in 2015–2017 to determine the risk of A. euteiches to these crops. Fields were preferentially chosen if they had a prior history of pea production, although very few dry bean fields were identified that had a pea or lentil crop in their rotation history. Root rot incidence and severity was very low in all crop types in all years (). As a result, there were very few root samples from these crops that showed any root rot symptoms, consequently there were very few samples retained for pathogen analysis. Aphanomyces euteiches was detected in a low number of alfalfa fields in 2015 and 2016, but no fields were positive in 2017. In 2016, 27% of dry bean fields were positive for A. euteiches, but a positive detection was always limited to one site per field, and root rot severity was always low. Soil baiting assays using pea or dry bean grown in soil collected from putative positive fields failed to confirm the presence of A. euteiches.
Table 6. Number of fields, mean root rot incidence (at damaging levels, DI > 3), severity (Sev), and per cent of fields positive for Aphanomyces euteiches (Ae), based on PCR assays from diseased root tissues, in Alberta in alfalfa, faba bean and dry bean fields surveyed in 2015–2017.
Discussion
Root rot was present in every pea field surveyed in all three provinces, with moderate to severe symptoms in 25–99% of fields assessed within each province. There were substantial differences among years but these differences can be attributed to sampling choice or time. Fields in Alberta in 2014 were selected for inclusion in the survey because they were severely diseased and therefore likely candidates for A. euteiches infestation. This approach skewed the results towards a high mean. In contrast, there were no differences in root rot incidence and severity among 2015–2017, when all fields were chosen arbitrarily based on crop district or county. In Saskatchewan in 2015, pea fields were assessed during pod fill in late July–early August, which was later in the growing season than other years, which would account for the higher levels of root rot observed in 2015.
This is the first study to demonstrate that A. euteiches occurs across the Prairie region, irrespective of soil type, and is also more severe on field pea compared with lentil. There was little or no difference in the incidence or severity of root rot or the proportion of pea fields positive for A. euteiches across soil zones. This was unexpected because precipitation in the black soil zone is generally higher and evapotranspiration lower than in the brown soil zone, with the dark brown zone being intermediate. Root rot, especially when caused by A. euteiches, was expected to be most frequent and most severe in regions with wetter soils, but this expectation was not supported upon examination of over 800 pea fields over four years. It is possible that localized precipitation events affected the distribution of A. euteiches, but this could not be measured due to the expansive areas surveyed.
Root rot was also present in every lentil field surveyed, based on assessment of over 200 fields in three years. However, severity was consistently and substantially lower than on field pea, especially in 2017, which was a very dry growing season in both Alberta and Saskatchewan. It is interesting to note that there were no differences in root rot level or frequency of A. euteiches associated with soil zone for the highly susceptible pea crop, but there were small but significant differences for the slightly less susceptible lentil crop. There were also fewer positive sites within lentil fields, whereas for the majority of positive pea fields, all sites within a field were positive. However, the proportion of positive sites within a field also varied by year, with fewer positive sites within a field in drier years compared with wet years. This substantiates the expectation that wetter conditions are more conducive for root rot, and especially for A. euteiches, but also indicates that root rot can be an important disease of pulse crops across all soil zones and environmental conditions.
The diagnostic component of the surveys relied solely on PCR techniques applied directly to root DNA, so that a large number of fields could be assessed quickly. Even then, only a proportion of sites and fields (samples with root rot severity > 3) were included so that a manageable number of samples could be assessed. A small number of fields in each province over 2 years were assessed by both classical isolation techniques (Esmaeili Taheri et al., Citation2017a) and the multiplex PCR assays. In general, PCR assays were positive at higher frequencies for all pathogens except F. acuminatum compared with isolation. Most importantly, isolation yielded very few cultures of A. euteiches because this pathogen grew slowly relative to other pathogens in the pea root rot complex, even on semi-selective media. The greatest success in obtaining A. euteiches was from samples collected early in the growing season (late May–early June) and from fields where early yellowing and stunting occurred (). This may indicate that infection of pea roots by A. euteiches occurs earlier than other pathogens. Difficulty in isolating A. euteiches was certainly one of the reasons why A. euteiches was not considered a major pathogen of pea in the Great Plains of North America, as previous surveys and identification relied on isolation from samples that were usually collected during flowering in July (Feng et al., Citation2010; Chittem et al., Citation2015). A systematic survey of the changes in pathogen composition in the roots over the growing season would aid in understanding pathogen dynamics leading to severe root rot.
Despite the high-throughput and rapid diagnosis capability of multiplex PCR, there were drawbacks to this technique. Results from the PCR assay for A. euteiches were sometimes difficult to score, and faint bands were usually scored as a negative reaction. Each A. euteiches PCR assay was repeated three times and, if there were discrepancies, the consensus of two reactions out of three was considered to be the true result. There was a trend towards a higher number of weak and inconsistent positive results in dry years, particularly in lentil. The results were interpreted conservatively to minimize false positives, but it is difficult to know if these weak bands were really positive samples with a low biomass in the roots, or true negatives. It was also difficult to interpret results for the F. avenaceum reaction because the primers for this species cross-reacted with F. acuminatum and F. tricinctum. This group can be considered as a species-complex, and thus it has been difficult to develop species-specific primers for individual species (Niessen et al., Citation2012). The ambiguity of scoring some of the PCR reactions forced us to consider F. avenaceum and F. acuminatum as a group, when they may be distinct species. Based on previous data from traditional culturing and isolation (Feng et al., Citation2010; Esmaeili Taheri et al., Citation2017a), and the PCR assays that gave clear results, we conclude that F. avenaceum is the dominant species, with F. acuminatum accounting for ~10% of positive reactions in this group. A series of multiplex PCR reactions to distinguish Fusarium species associated with pea root rot was recently published (Zitnick-Anderson et al., Citation2018), but this methodology was not available at the time when surveys and analysis were conducted. It would be useful to test this methodology on archived DNA samples to determine if this protocol improves species delineation.
Finally, the pea root samples used for DNA extraction were not subject to a surface disinfestation step prior to DNA extraction, whereas isolation techniques almost always commence with this step. This could thus also account for the higher frequencies of detection observed with the PCR technique, as many soil saprophytes that colonize the root surface and/or dead tissues would have been included in the detections.
Although a number of Fusarium species were frequently detected in pea and lentil roots, previous pathogenicity tests with many of the species detected indicated that F. avenaceum and F. solani were the most aggressive on pea, while lentil was most susceptible to F. avenaceum and less so to F. solani (Safarieskandari et al., Citation2016; Willsey et al., Citation2018). Although common, F. redolens and F. oxysporum caused only mild root rot symptoms on pea and lentil (Safarieskandari et al., Citation2016). We conclude that, in addition to A. euteiches, F. avenaceum and F. solani were the major causal agents of pea root rot on the Canadian prairies. Across all years and locations, 55% and 43% of pea samples and fields, and 73% and 59% of lentil samples and fields were positive for F. avenaceum and/or F. solani (i.e. A. euteiches was not present in these samples and fields). Furthermore, in fields where A. euteiches was present, it almost always occurred as a complex with these same Fusarium spp., as only 1.6% of lentil samples and 6% of pea samples had A. euteiches in the absence of F. avenaceum and/or F. solani. Fusarium redolens was also commonly detected, and can have a synergistic relationship with A. euteiches and other Fusarium spp. to increase root rot severity (Willsey et al., Citation2018). Fusarium graminearum, the primary causal agent of fusarium head blight of wheat in the Canadian prairies, was infrequently detected in pea and lentil fields in Alberta but was detected in 5–32% of fields in Manitoba and Saskatchewan. Fusarium graminearum has recently spread westward in Alberta (Harding et al., Citation2018), but has been an issue in wheat fields in Saskatchewan and Manitoba for a number of years (Clear et al., Citation2005). This result supports previous reports that F. graminearum can be isolated from pea roots and can cause root rot on pea and lentil (Chongo et al., Citation2001; Fernandez, Citation2007).
Rhizoctonia solani was also frequently detected in pea roots, but was rarely isolated in the study by Esmaeili Taheri et al. (Citation2017a). However, it is a common soil-borne pathogen and saprophyte, and was frequently isolated from roots and crowns of pea and other hosts in previous studies (Melzer et al., Citation2016; Yu et al., Citation2018). There is substantial diversity in the pathogenicity of R. solani, with only specific anastomosis groups (AG) able to infect pea and lentil. The PCR primers used in this study targeted all AGs of R. solani, and thus could not differentiate between saprophytic and pathogenic strains. However, the root rot symptomatology observed was rarely characteristic of R. solani. While it is likely that R. solani is involved in the root rot complex, either as a later colonizer of decaying root tissue and/or by acting synergistically with other members of the complex, our results only weakly confirm the role of this pathogen.
This is also the likely scenario for Pythium spp. Like R. solani, and Fusarium spp., Pythium is a common soil-borne inhabitant and was the predominant oomycete genus in pea rhizosphere soil (Esmaeili Taheri et al., Citation2017b). It should be noted that the common practice in the prairies is to treat pea and lentil seeds with synthetic seed treatments active against Pythium, Fusarium and Rhizoctonia to prevent pre-emergent and post-emergent damping-off. Aphanomyces euteiches has a wide host range within the legume family (Moussart et al., Citation2008), and with its widespread occurrence in the prairies there was concern that it could also affect the nascent faba bean industry across the region and the dry bean industry in southern Alberta and Manitoba. Alfalfa is also susceptible to A. euteiches (Malvick & Grau, Citation2001). Therefore, surveys were conducted in these crops to assess the risk of A. euteiches. Dry bean in southern Alberta is grown under irrigation, so is rarely grown in a rotation that includes pea or lentil because these crops are not typically irrigated. However, faba bean is replacing pea in many of the wetter portions of the area previously used for pea production, which have suffered from yield loss due to pea root rot. Aphanomyces euteiches was not detected in any faba bean fields, was rarely detected in alfalfa and dry bean fields, and none of these crops displayed high levels of root rot. Since A. euteiches had not previously been reported on dry bean in Alberta, further efforts to isolate A. euteiches from dry bean fields that had tested positive were initiated. However, soil baiting assays failed to produce A. euteiches. It appears that A. euteiches currently poses little risk to other legume crops commonly grown on the prairies other than pea and lentil. Nevertheless, periodic surveys that include a pathogen diagnostic component should be performed to identify if this risk changes over time.
In conclusion, this extensive multi-year survey is the first to demonstrate that A. euteiches is widespread on pea and lentil across the Canadian prairies, even though it was only first reported in Saskatchewan in 2012 and Alberta in 2013. Complete yield loss associated with severe root rot was observed in many fields, particularly in the wet growing seasons of 2014 and 2016. Although A. euteiches poses a high risk to pea production and is considered the most damaging pea pathogen worldwide (Gaulin et al., Citation2007), F. avenaceum and F. solani are also major contributors to the root rot problem independently and more so when both Fusarium spp. and A. euteiches are both present. The timing of infection and the role each contributes to disease development and symptom progression throughout the season has not been elucidated. These surveys documented that there is a large complex of soil-borne pathogens and/or saprophytes associated with pea root rot, warranting further evaluation of the impact of these pathogens on yield loss. Finally, our results indicated that wet or dry weather conditions are key indicators of root rot severity, particularly for lentil. Wetter conditions are more conducive for root rot, and especially for A. euteiches, but root rot of pulse crops frequently occurs even in dry regions.
Supplementary Tables 1 and 2
Download MS Word (17.7 KB)Acknowledgements
The authors thank H. Andkhoie, K. Bassendowski, D.A. Burke, G.C. Daniels, T. Dubitz, C. Mueller, C. Vucurevich and K. Wetzel for technical assistance. There was a large network of Saskatchewan Ministry of Agriculture staff that surveyed pea and lentil crops in Saskatchewan and we gratefully acknowledge their assistance. We thank all producers that co-operated with surveillance efforts and field sampling. Funding for this project was from the Pulse Science Cluster of Agriculture and Agri-Food Canada and Pulse Canada, Alberta and Saskatchewan Pulse Growers, and the Alberta Crop Industry Development Fund.
Supplementary material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2018.1547792.
Additional information
Funding
References
- Armstrong-Cho C, Tetreault M, Banniza S, Bhadauria V, Morrall RAA. 2014. Reports of Aphanomyces euteiches in Saskatchewan. Can Plant Dis Surv. 94:193–194.
- Banniza S, Bhadauria V, Peluola CO, Armstrong-Cho C, Morrall RAA. 2013. First report of Aphanomyces euteiches in Saskatchewan. Can Plant Dis Surv. 93:163–164.
- Bilgi VN, Bradley CA, Khot SD, Grafton KF, Rasmussen JB. 2008. Response of dry bean genotypes to Fusarium root rot, caused by Fusarium solani f. sp. phaseoli, under field and controlled conditions. Plant Dis. 92:1197–1200.
- Bogale M, Wingfield BD, Wingfield MJ, Steenkamp ET. 2007. Species-specific primers for Fusarium redolens and a PCR-RFLP technique to distinguish among three clades of Fusarium oxysporum. FEMS Microbiol Lett. 271:27–32.
- Bowley SR. 2015. A hitchhiker’s guide to statistics in biology: generalized linear mixed model edition. Kincardine (ON): Plants et al., Inc.
- Chatterton S, Bowness RT, Harding MW. 2015a. First report of root rot of field pea caused by Aphanomyces euteiches in Alberta, Canada. Plant Dis. 99:288.
- Chatterton S, Harding M, Bowness R, Strydhorst S, Cleland C, Storozynsky Q, Dubitz T, Nielsen J, Olson M. 2015b. Survey of root rot in Alberta field pea in 2014. Can Plant Dis Surv. 95:170–172.
- Chittem K, Mathew F, Gregoire M, Lamppa R, Chang Y, Markell S, Bradley C, Barasubiye T, Goswami R. 2015. Identification and characterization of Fusarium spp. associated with root rots of field pea in North Dakota. Eur J Plant Pathol. 143:641–649.
- Chongo G, Gossen BD, Kutcher HR, Gilbert J, Turkington TK, Fernandez MR, McLaren D. 2001. Reaction of seedling roots of 14 crop species to Fusarium graminearum from wheat heads. Can J Plant Pathol. 23:132–137.
- Clear RM, Patrick SK, Gaba D, Abramson D, Smith DM. 2005. Prevalence of fungi and fusariotoxins on hard red spring and amber durum wheat seed from western Canada, 2000 to 2002. Can J Plant Pathol. 27:528–540.
- Doohan FM, Parry DW, Jenkinson P, Nicholson P. 1998. The use of species-specific PCR-based assays to analyse Fusarium ear blight of wheat. Plant Pathol. 47:197–205.
- Esmaeili Taheri A, Chatterton S, Foroud NA, Gossen BD, McLaren DL. 2017a. Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. Eur J Plant Pathol. 147:489–500.
- Esmaeili Taheri A, Chatterton S, Gossen BD, McLaren DL. 2017b. Degenerate ITS7 primer enhances oomycete community coverage and PCR sensitivity to Aphanomyces species, economically important plant pathogens. Can J Microbiol. 63:769–779.
- Etebu E, Osborn AM. 2009. Molecular assays reveal the presence and diversity of genes encoding pea footrot pathogenicity determinants in Nectria haematococca and in agricultural soils. J Appl Microbiol. 106:1629–1639.
- Feng J, Hwang R, Chang KF, Hwang SF, Strelkov SE, Gossen BD, Conner RL, Turnbull GD. 2010. Genetic variation in Fusarium avenaceum causing root rot on field pea. Plant Pathol. 59:845–852.
- Fernandez MR. 2007. Fusarium populations in roots of oilseed and pulse crops grown in eastern Saskatchewan. Can J Plant Sci. 87:945–952.
- Gangneux C, Cannesan MA, Bressan M, Castel L, Moussart A, Vicré-Gibouin M, Driouich A, Trinsoutrot-Gattin I, Laval K. 2014. A sensitive assay for rapid detection and quantification of Aphanomyces euteiches in soil. Phytopathology. 104:1138–1147.
- Gaulin E, Jacquet C, Bottin A, Dumas B. 2007. Root rot disease of legumes caused by Aphanomyces euteiches. Mol Plant Pathol. 8:539–548.
- Geiser DM, Del Mar Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. 2004. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 110:473–799.
- Gossen B, Conner R, Chang K-F, Pasche JS, McLaren DL, Henriquez MA, Chatterton S, Hwang S-F. 2016. Identifying and managing root rot of pulses on the northern Great Plains. Plant Dis. 100:1965–1978.
- Harding MW, Howard RJ, Feng J, Laflamme P, Turkington TK, Grafenhan T, Daniels GC. 2018. Monitoring Fusarium graminearum in Alberta: looking back 20 years. Can J Plant Pathol. 40:141.
- Hwang SF, Gossen BD, Conner RL, Chang KF, Turnbull GD, Lopetinsky K, Howard RJ. 2007. Management strategies to reduce losses caused by rhizoctonia seedling blight of field pea. Can J Plant Sci. 87:145–155.
- Infantino A, Kharrat M, Riccioni L, Coyne CJ, McPhee KE, Grünwald NJ. 2006. Screening techniques and sources of resistance to root diseases in cool season food legumes. Euphytica. 147:201–221.
- Jiménez-Fernández D, Montes-Borrego M, Navas-Cortés JA, Jiménez-Díaz RM, Landa BB. 2010. Identification and quantification of Fusarium oxysporum in planta and soil by means of an improved specific and quantitative PCR assay. Appl Soil Ecol. 46:372–382.
- Lievens B, Brouwer M, Vanachter ACRC, Cammue BPA, Thomma BPHJ. 2006. Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 171:155–165.
- Malvick DK, Grau CR. 2001. Characteristics and frequency of Aphanomyces euteiches races 1 and 2 associated with alfalfa in the midwestern United States. Plant Dis. 85:740–744.
- McRoberts N, Hughes G, Madden LV. 2003. The theoretical basis and practical application of relationships between different disease intensity measurements in plants. Ann Appl Biol. 142:191–211.
- Melzer MS, Yu H, Labun T, Dickson A, Boland GJ. 2016. Characterization and pathogenicity of Rhizoctonia spp. from field crops in Canada. Can J Plant Pathol. 38:367–374.
- Mishra PK, Fox RTV, Culham A. 2003. Development of a PCR-based assay for rapid and reliable identification of pathogenic Fusaria. FEMS Microbiol Lett. 218:329–332.
- Moussart A, Even MN, Tivoli B. 2008. Reaction of genotypes from several species of grain and forage legumes to infection with a French pea isolate of the oomycete Aphanomyces euteiches. Eur J Plant Pathol. 122:321–333.
- Nicholson P, Simpson D, Weston G, Rezanoor H, Lees A, Parry D, Joyce D. 1998. Detection and quantification of Fusarium culmorum and Fusarium graminearum in cereals using PCR assays. Physiol Mol Plant Pathol. 53:17–37.
- Niessen L, Gräfenhan T, Vogel RF. 2012. ATP citrate lyase 1 (acl1) gene-based loop-mediated amplification assay for the detection of the Fusarium tricinctum species complex in pure cultures and in cereal samples. Int J Food Microbiol. 158:171–185.
- O’Brien HE, Parrent JL, Jackson JA, Moncalvo J-M, Vilgalys R. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 71:5544–5550.
- Pfender W, Delwiche P, Grau C, Hagedorn D. 1984. A medium to enhance recovery of Aphanomyces from infected plant tissue. Plant Dis. 68:845–847.
- Pfender WF, Hagedorn DJ. 1983. Disease progress and yield loss in Aphanomyces root rot of peas. Phytopathology. 73:1109–1113.
- Safarieskandari S, Chatterton S, Hall LM. 2016. Evaluation of host range of Fusarium species from pea fields in Alberta. Can J Plant Pathol. 38:268.
- Schroeder KL, Okubara PA, Tambong JT, Lévesque CA, Paulitz TC. 2006. Identification and quantification of pathogenic Pythium spp. from soils in eastern Washington using real-time polymerase chain reaction. Phytopathology. 96:637–647.
- Statistics Canada. 2017. Table 001-0010 - Estimated areas, yield, production and average farm price of principal field crops, in metric units, annual, CANSIM (database). [ accessed 2017 04-18]. http://www5.statcan.gc.ca/cansim/a26?lang=eng&id=10010.
- Turner AS, Lees AK, Rezanoor HN, Nicholson P. 1998. Refinement of PCR-detection of Fusarium avenaceum and evidence from DNA marker studies for phenetic relatedness to Fusarium tricinctum. Plant Pathol. 47:278–288.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: aGuide toMethods and Applications. Vol. 1990. San Diego (CA): Academic Press; p. 315–322.
- Williams KJ, Dennis JI, Smyl C, Wallwork H. 2002. The application of species-specific assays based on the polymerase chain reaction to analyse Fusarium crown rot of durum wheat. Australasian Plant Pathol. 31:119–127.
- Willsey TL, Chatterton S, Heynen M, Erickson A. 2018. Detection of interactions between the pea root rot pathogens Aphanomyces euteiches and Fusarium spp. using a multiplex qPCR assay. Plant Pathol. 67:1912–1923.
- Yu HT, Zhou Q, Fu HT, Chang KF, Hwang SF, Strelkov SE. 2018. Pathogenicity and genetic diversity of Rhizoctonia solani isolates from field pea and other crops in Alberta, Canada. Can J Plant Pathol. 40:144.
- Zitnick-Anderson K, Simons K, Pasche JS. 2018. Detection and qPCR quantification of seven Fusarium species associated with the root rot complex in field pea. Can J Plant Pathol. 40:261–271.
