Abstract
The biocontrol strain BJ-1 was isolated from a contaminated Magnaporthe oryzae culture plate and identified as Bacillus subtilis based on morphological characteristics, Biolog Microlog identification and 16S rDNA sequence analysis. The dual culture test showed that BJ-1 has a wide antimicrobial spectrum, and is especially potent against M. oryzae. The culture filtrates of BJ-1 significantly inhibited mycelial growth of M. oryzae P131, caused mycelial disintegration and induced swollen germ tubes. RT-PCR revealed the expressions of seven lipopeptide synthesis genes, encoding surfactin (srfAA and SPAB-ERIB), fengycin (fenB and FENCEA), subtilin (spa and spaS) and bacilysin (BACD) in BJ-1. Application of 1 × 108 CFU mL−1 bacterial cells or 5% culture filtrates of BJ-1 completely suppressed rice blast on detached leaves. In pot experiments, both spray and seed treatments using culture broth provided more than 50% disease suppression on rice blast. Interestingly, rice seeds soaked in culture broth of BJ-1 before planting induced host systemic resistance to inhibit rice blast development and promote plant growth, with disease reduction of over 70%, at a similar level as the fungicide tricyclazole. Expressions of two PR genes (OsPR1a and OsPR10) and three genes (OsPAL, OsICS1 and OsAOS2) known to function in salicylate- and jasmonate-dependent defence signalling pathways in rice plants after seed treatment by culture broth of BJ-1 were significantly induced. This is the first description of the use of B. subtilis as a seed treatment for control of rice blast and these results indicate that B. subtilis BJ-1 is a highly promising biocontrol agent for suppression of rice blast disease.
Résumé
La souche BJ-1 pour la lutte biologique a été isolée à partir d’une boîte de culture de Magnaporthe oryzae contaminée et a été identifiée en tant que Bacillus subtilis en se basant sur les caractéristiques morphologiques, le système d’identification MicroLog de Biolog et l’analyse de la séquence 16S de l’ADNr. Le test de double culture a montré que la souche BJ-1 possède un large spectre antimicrobien et qu’elle est particulièrement efficace contre M. oryzae. Les perméats résultant de la culture de BJ-1 ont significativement inhibé la croissance mycélienne de M. oryzae P131, ont causé la désintégration du mycélium et ont induit le gonflement des filaments germinatifs. La RT-PCR a révélé l’expression de sept gènes de synthèse de lipopeptides codant la surfactine (srfAA et SPAB-ERIB), la fengycine (fenB et FENCEA), la subtiline (spa et spaS) et la bacilysine (BACD) chez BJ-1. L’application de 1 × 108 CFU/ml de cellules bactériennes ou 5% de perméats de cultures de BJ-1 a entièrement éliminé la pyriculariose de feuilles détachées. Lors d’expériences en pots, la pulvérisation et le traitement des semences avec un bouillon de culture ont permis d’éliminer plus de 50% de la pyriculariose. Chose intéressante, le trempage de la semence de riz dans le bouillon de culture de BJ-1 avant la plantation a induit la résistance systémique de l’hôte, ce qui a inhibé le développement de la pyriculariose et favorisé la croissance des plants, avec comme résultat une réduction de plus de 70% de la maladie, ce qui est analogue au résultat obtenu avec le tricyclazole. L’expression de deux gènes PR (OsPR1a et OsPR10) et de trois autres (OsPAL, OsICS1 et OsAOS2), impliqués dans les voies de signalisation de défense dépendantes du salicylate et du jasmonate des plants de riz, a été significativement induite après avoir traité les semences avec le bouillon de culture de BJ-1. Il s’agit de la première description de l’utilisation de B. subtilis en tant que traitement pour semences dans le but de lutter contre la pyriculariose. En outre, ces résultats indiquent que la souche BJ-1 à base de B. subtilis est un agent de lutte biologique fort prometteur en ce qui a trait à la suppression de la pyriculariose.
Introduction
Rice blast caused by Magnaporthe oryzae is one of the most destructive diseases on rice worldwide. The disease has been found in more than 85 countries and causes 30% annual yield losses on average (Dagdas et al., Citation2012). Due to the complex genetic background of M. oryzae and widespread use of cultivars with single gene resistance, it is difficult to maintain durable resistance against rice blast; hence, chemical fungicides are the most common method for disease control. However, chemical fungicides can lead to non-target toxicity, environmental pollution, biodiversity reduction and many other disadvantages which are hard to overcome (Hirooka & Ishii, Citation2013). Microorganisms and their metabolites have attracted attention as potential biocontrol agents to reduce fungicide use (Li et al., Citation2006, Citation2011; Kim et al., Citation2009).
Fig. 1 Neighbor-Joining tree showing the position of B. subtilis BJ-1 compared with related organisms in a 16S rDNA gene tree. GenBank accession numbers are provided. Numbers at nodes indicate per cent bootstrap support based on 1000 replicates, and only those branches with greater than 50% bootstrap support were labelled. The scale bar at the bottom indicated genetic distance units based on Nei’s genetic distance.
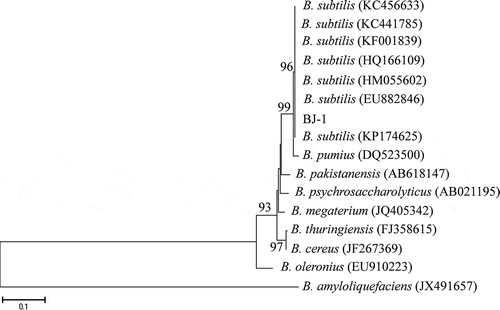
Fig. 2 Identification and expression of antifungal genes from B. subtilis BJ-1. a, PCR amplification of lipopeptide biosynthetic genes from genomic DNA of BJ-1. b, Expression levels of lipopeptide biosynthetic genes were detected using RT-PCR. The housekeeping gene rpoB was amplified as control. Marker: TaKaRa DL1000 (TaKaRa, Dalian, China).
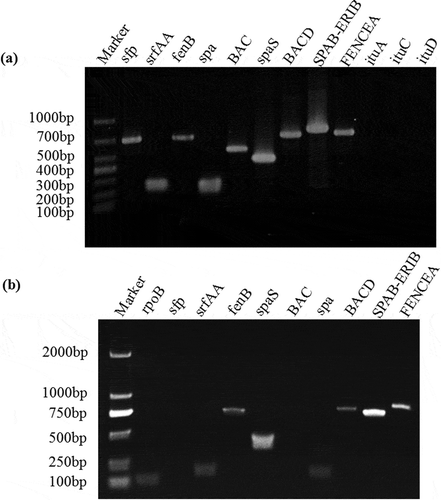
Fig. 3 (Colour online) Effects of culture filtrates of B. subtilis BJ-1 on mycelial morphology, conidial germination and appressorial formation of M. oryzae strain P131. a, Mycelia of M. oryzae without culture filtrate of BJ-1 (CK). b, Mycelia of M. oryzae treated with 0.5% culture filtrate of BJ-1. Arrows indicate abnormal hyphal fragments. c, Conidial germination and appressorial formation without culture filtrate of BJ-1 (CK). d, Malformation of germ tubes with 0.5% culture filtrate of BJ-1. Arrows indicate bulbous germ tubes. Bar = 10 µm.
Fig. 4 (Colour online) Control of rice blast disease on detached rice leaves by various concentrations of culture broth (culture filtrate plus bacterial cells), culture filtrates and B. subtilis BJ-1 bacterial cells. a–f, The culture broth of BJ-1 was added to conidial suspensions to obtain final concentrations of 0, 0.5%, 1%, 2.5%, 5% or 10%. g–l, The culture filtrates of BJ-1 were added to conidial suspensions to obtain final concentrations of 0, 0.5%, 1%, 2.5%, 5% or 10%. m–p, BJ-1 bacterial cells obtained by centrifugation were washed three times, resuspended in distilled water and mixed with conidial suspensions to obtain a final concentration of 0, 1 × 107, 1 × 108 or 1 × 109 CFU mL−1. Conidial suspensions of M. oryzae were used at 5 × 105 conidia mL−1.
Fig. 5 (Colour online) Control of rice blast in greenhouse by culture broth of B. subtilis BJ-1 on five-leaf stage rice seedlings. a, Rice seedlings without M. oryzae P131 inoculation and BJ-1 treatment served as blank control. b, Rice seedlings only sprayed with conidial suspension (5 × 105 conidia mL−1) served as negative control. c, Rice seedlings sprayed with culture broth of BJ-1 mixed conidial suspension to obtain a final concentration of 10% (v/v). d, Rice seedlings sprayed with tricyclazole (750 μg mL−1) mixed conidial suspension as positive control. e, Rice seeds were soaked with culture broth (10%, v/v) of BJ-1 for 24 h and planted, and then conidial suspension (5 × 105 conidia mL−1) was sprayed onto rice seedlings. f, Seed treatment promoted plant growth of rice. g, Rice plants without any treatments.

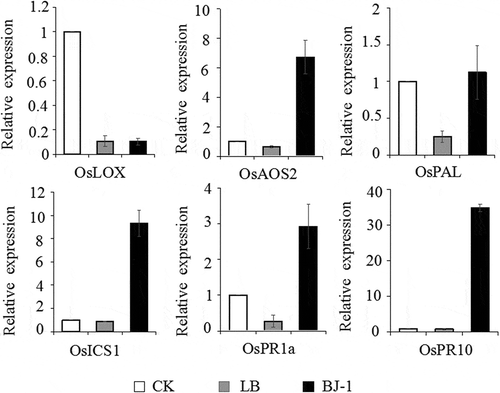
Fig. 6 Expression analysis by quantitative RT-PCR of defence-related genes in five-leaf stage rice seedlings after seeds were soaked with culture broth (10%, v/v) of BJ-1. Rice seedlings after seeds were treated with water and LB medium as blank control and negative control, respectively. Expression of OsACTIN gene was used as an internal control. Relative abundance of transcripts of defence-related genes was normalized by comparing with the expression of these genes in rice seedlings after seeds were treated with water (relative transcript level = 1). Data were collected from three independent experiments with three replicates in each treatment. Vertical bars indicated standard error of measurements for three sets of experiments.
Various naturally occurring microorganisms are reported to inhibit plant pathogenic fungi. Among these, B. subtilis was shown to produce a wide variety of antimicrobial substances and enzymes, which inhibit or kill phytopathogens (US Environmental Protection Agency, Citation1997; Leelasuphakul et al., Citation2006; Sajitha & Dev, Citation2016). The bacterium produces novel antibiotics such as difficidin and oxydifficidin that have activity against a wide spectrum of aerobic and anaerobic bacteria, as well as more common antibiotics such as bacitracin, bacillin and bacillomycin B (US Environmental Protection Agency, Citation1997). A variety of strains of Bacillus and particularly B. subtilis are currently commercialized as bio-fungicides and numerous studies support the great potential of such strains in controlling multiple diseases on a wide range of host plant species (Cawoy et al., Citation2011). Moreover, B. subtilis may promote plant growth and induce host systemic resistance (Kloepper et al., Citation2004; Sha et al., Citation2016). An antagonistic strain of B. subtilis UMAF6639 conferred protection to melon plants against cucurbit powdery mildew by activation of jasmonate and salicylic acid-dependent defence responses (Garcia-Gutiérrez et al., Citation2013). Seed treatment with B. subtilis effectively suppressed Rhizoctonia solani on peanut (Turner & Backman, Citation1991) and tomato (Zohora et al., Citation2016). However, the use of seed treatment of rice seeds with B. subtilis to acquire induced host systemic resistance against rice blast has not been previously reported.
We found a contaminant, named BJ-1, in a culture of M. oryzae which caused the formation of a zone of inhibition. The objectives of this study were to: (1) identify isolate BJ-1; (2) evaluate the inhibition characteristics of BJ-1 against M. oryzae and confirm the level of disease suppression of rice blast; and (3) assess the biocontrol potential of BJ-1 as a seed treatment agent.
Materials and methods
Microorganisms, media and plant materials
Strain BJ-1 was obtained from a contaminated oatmeal-tomato agar (OTA) plate of M. oryzae in the Key Lab of Plant Pathology of Hubei Province, Huazhong Agricultural University, Wuhan, China in 2015. It was purified and cultured onto solid Luria-Bertani (LB) plates and preserved in 25% glycerol at −80°C for further study. To prepare culture filtrates, submerged cultures were grown at 37°C on a rotary shaker at 180 rpm in 250 mL flasks filled with 100 mL of liquid LB medium. After incubation for 3 days, the culture broth was centrifuged (8000 rpm, 20 min). The decanted cell-free supernatant was passed through a 0.22-μm pore size filter to exclude bacteria, and the culture broth and the cell-free culture filtrates were both used to test for antifungal and disease suppression activity in the subsequent bioassays.
Preparation of conidial suspensions of M. oryzae followed Li et al. (Citation2011), and is briefly summarized below. Mycelial plugs were inoculated onto OTA at 28°C and cultured for 7 d, and then fungal mycelia were scraped off the surface and washed off. After the agar surface had dried in a flow hood, it was covered with two layers of autoclaved wet cheesecloth and incubated at 28°C under continuous light for 48 h to induce sporulation. Conidial suspensions of M. oryzae were filtered through three layers of cheesecloth to remove mycelial fragments and agar pieces, adjusted to 5 × 105 conidia mL−1, and used in bioassays. For fungal inhibition tests, 12 plant pathogens from our culture collection () were grown on potato dextrose agar (PDA) at 25°C for up to 7 d prior to use as mycelial plugs.
Table 1. Antagonism of B. subtilis BJ-1 toward selected plant pathogenic fungi in dual culture test on PDA plates.
Rice ‘Fengliangyouxiang-1' (Hefei Fengle Seed Co., China), which is known to be moderately susceptible to rice blast, was used in inoculation tests. Rice seeds were surface-sterilized with potassium permanganate solution (0.1%) for 12 h and rinsed with sterile distilled water. Seeds were then incubated on moist absorbent paper at 37°C. After 2 days, 90 germinated seeds were transplanted into each of 15 (20 cm × 25 cm × 30 cm) plastic pots containing field soil (University rice field, Wuhan, China; paddy soil, pH 6.0–6.5). The potted plants were grown in a greenhouse at 24–30°C with natural sunlight and irrigated with tap water as needed. Seedlings at the five-leaf stage were used for further bioassays.
Identification of strain BJ-1
The characteristics of strain BJ-1 including colony morphology, Gram staining and endospore staining were determined. Cultures were streaked onto LB agar plates and incubated in a 37°C incubator for 24 h, and then colony morphology and transparency were recorded. Crystal violet was used in Gram staining, and purple staining was positive while pink staining was negative. Malachite-green method was used for endospore detection (Reynolds et al., Citation2009).
The bacterial identification system, GEN III MicroPlate™ test panel (Biolog, Hayward, CA, USA) which is based on carbon metabolism was conducted on strain BJ-1 following the manufacturer’s instructions. Briefly, cultures were grown on Biolog BUG agar, and cells from each plate was removed with a sterile swab, re-suspended in Biolog dilution medium, and adjusted to ~20% transmittance. The bacterial suspension was pipetted into a Biolog Gram-positive 96-well microplate and incubated at 35°C. The microplate was read at 4–6 h and again at 16–24 h using a microplate reader, and the Microlog Gram-positive database was used to yield a carbon utilization metabolic fingerprint for bacterial identification. Identification confidence was evaluated based on per cent probability (PROB), similarity index (SI) and distance (DIST) after 16–24 h incubation.
Molecular identification of strain BJ-1 was conducted using 16S rDNA sequence analysis. Cultures were grown in 2 mL of LB liquid medium and shaken at 37°C for 24 h. Genomic DNA was extracted using a TIANamp Bacteria DNA kit based on the manufacturer’s instructions (Tiangen Biotech, Beijing, China). The 16S rDNA of strain BJ-1 was amplified with primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′- GGTTACCTTGTTACGACTT −3′) following Kim et al. (Citation2009). The PCR products were purified using E.Z.N.A. Cycle Pure Kit (OMEGA Bio-Tek, Guangzhou, China), and sent to Wuhan Genecreat Biological Engineering Co. (Wuhan, China) for sequencing. A consensus sequence of the forward and reverse reads was generated with DNAMAN (version 8.0). The consensus sequence was compared with GenBank, and the highest matching sequences were downloaded and aligned using ClustalX (version 2.0) (Thompson et al., Citation2002). The Neighbour-Joining algorithm was used in MEGA5.0 to construct a phylogenetic tree with 1000 bootstrap replicates (Li et al., Citation2011).
All experiments of morphological observation, Biolog identification and 16S rDNA sequence analysis were conducted at least twice.
Antagonism of BJ-1 to various plant pathogenic fungi
To evaluate the antifungal activity of strain BJ-1, the dual culture method was used (Pedersen et al., Citation2010) with a range of phytopathogenic fungi (). Cultures were grown in LB broth and shaken at 180 rpm for 3 days in a 37°C incubator. A 10 μL aliquot of bacterial suspension was placed in a straight line down the centre of each PDA plate, and replicate hyphal agar plugs were placed on each side of the line at a 35 mm distance. Separate plates were made for each of the 12 phytopathogenic fungi plus a non-inoculated LB broth control. After the plates were almost fully grown, the distance between colonies was measured. For each treatment, mean value from three plates served as a replicate and the experiment was repeated three times.
Identification and expression of antifungal genes from BJ-1
Presence of genes corresponding to various known antifungal compounds in genomic DNA of BJ-1 were first verified. Genomic DNA was extracted using TIANamp Bacteria DNA kit (Tiangen Biotech, Beijing, China) and amplification using specific primers of antifungal lipopeptide biosynthetic genes for iturin, fengycin, surfactin, bacillomycin, mycosubtilin and bacilysin was conducted (Yang et al., Citation2015, Table S1). PCR conditions were as follows: 5 min of denaturation at 94°C, followed by 30 cycles of 94°C for 30 s, 52 to 62°C for 30 s (depending on primer pair, Table S1), and 72°C for 30 s to 2 min, and a final extension at 72°C for 10 min. The experiment was repeated three times.
Expression of antifungal genes was further analysed using RT-PCR. Total RNA was extracted with the OMEGA Bio-tek Bacteria DNA Kit (OMEGA, Guangzhou, China). Reverse transcription assays were performed using TransScriptTM One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). For RT-PCR, total RNA was collected from bacteria grown for 3 days in liquid LB. RT-PCR mixtures were composed of 12.5 μL of 2× HieffTM PCR Master Mix (YeSen, Shanghai, China), 1 μL of cDNA, 1 μL of each primer, and nuclease-free water to a final volume of 25 μL. The housekeeping gene rpoB was amplified as a control for RT-PCR (Caamaño-Antelo et al., Citation2015). Primer pairs for B. subtilis genes were used to detect the gene expression (Table S1). The experiment was repeated three times.
Suppression of mycelial growth of M. oryzae by culture filtrates of BJ-1
The effects of culture filtrates on suppression of mycelial growth of M. oryzae strain P131 were assessed. The filtrates from 3-day-old LB broth cultures of strain BJ-1 were incorporated into potato dextrose broth (PDB) at final concentrations of 0.5, 1, 2.5, 5, 10 or 20% (v/v), while LB broth without BJ-1 was added into PDA as a control. A 5-mm-diameter mycelial plug of P131 from a 7-day-old PDA culture was inoculated into each flask. After incubation in shake culture (180 rpm) for 7 d at 28°C, mycelial mats were passed through pre-weighed filter paper (Chongqing Chuandong Chemical, Co. Ltd, Chongqing, China), dried at 60°C and weighed. For each treatment, mean values from three plates served as a replicate and the experiment was repeated three times.
Effects of culture filtrates of BJ-1 on conidial germination and appressorial formation of M. oryzae in vitro
Conidial suspensions (final concentration 5 × 105 conidia mL−1) containing 0.025% Tween 20 were prepared and mixed with the culture filtrates of BJ-1 to final concentrations of 0.5, 1, 2.5, 5, 10 or 20% (v/v). Conidial suspension containing Tween 20 mixed with LB broth was used as a control. Three 5-μL droplets of each mixture were placed onto coverslips which were placed into Petri dishes lined with moist absorbent paper. All plates were incubated at 28°C in darkness with 100% relative humidity in a growth chamber. After 16 h, conidial germination and appressorial formation rates were assessed. For each replicate, at least 300 spores from three replicate plates were counted to calculate mean value. The experiment was conducted three times.
Effect of BJ-1 on rice blast development on detached rice leaves
Strain BJ-1 was grown in LB (200 mL) in 250 mL flasks at 37°C and shaken at 180 rpm for 3 d. Washed cells were then centrifuged and mixed into conidial suspensions of P131 (final concentration 5 × 105 conidia mL−1), and bacterial concentrations were adjusted to 107, 108 or 109 CFU mL−1 with a dilute 0.025% Tween 20 solution. Distilled water mixed into conidial suspensions with Tween 20 solution served as a control. At the same time, 3-day-old culture broth or culture filtrates were diluted to final concentrations of 0.5, 1, 2.5, 5 and 10% (v/v), and mixed with conidial suspensions of P131 to 5 × 105 conidia mL−1 in 0.025% Tween 20. Conidial suspensions containing Tween 20 mixed with LB broth was used as a control.
Detached leaves of rice at the five-leaf stage were placed into dishes lined with moist absorbent paper, and each leaf was inoculated with three 5-μL droplets of the mixtures prepared above. All dishes were incubated at 28°C with 100% relative humidity, first in darkness for 30 h, and then in constant light for 2 d. Disease incidence and lesion area were measured from 12 leaves in each treatment, and the experiments were repeated three times.
Effect of BJ-1 on rice blast development in pot experiments
Rice seedlings at the five-leaf stage were treated as follows with 30 mL per pot: (1) blank check without M. oryzae P131 inoculation nor BJ-1 treatment; (2) negative control sprayed with a mixture of LB medium and conidial suspension (5 × 105 conidia mL−1) of P131 amended with 0.025% Tween 20; (3) positive control sprayed with a mixture of the fungicide tricyclazole (750 μg mL−1) and conidial suspension amended with 0.025% Tween 20; and (4) treatment sprayed with a mixture of culture broth from 3-day-old cultures (10%, v/v) and conidial suspension amended with 0.025% Tween 20. In addition, a fifth treatment (5) consisted of rice seeds germinated in 3-day-old culture broth of strain BJ-1 (10%, v/v) for 24 h, and then were planted after rinsing with water. At the five leaf stage, seedlings were sprayed with conidial suspensions.
All treated rice seedlings were incubated in a growth chamber with 100% relative humidity at 28°C in the dark for 30 h and then in constant light for 3 d and subsequently moved to a 28 ± 3°C greenhouse. Disease severity at 6 d was evaluated following Prabavathy et al. (Citation2006) on a 0–9 scale. Three pots (20 cm × 25 cm × 30 cm) of rice seedlings were used for each treatment. In each pot, 30 seedlings were chosen randomly for evaluation of disease incidence and severity using the scale mentioned above, and the experiments were conducted three times.
Expression of defence-related genes in rice plants after seed treatment with BJ-1
Rice seeds were immersed in a 3-day-old culture broth of strain BJ-1 (10%, v/v) for 24 h, and then were planted after rinsing with water. Seeds treated with water and LB medium were regarded as blank control and negative control, respectively. All plants were incubated in a growth chamber with the conditions described previously. Three pots of rice seedlings were used for each treatment as three replicates. At the five leaf stage, five seedlings in each pot were sampled randomly for RNA extraction. Total RNA was extracted from leaves using the Takara MiniBEST Universal RNA Extraction Kit (TaKaRa, Dalian, China). Reverse transcription assays were performed using TransScriptTM One-Step gDNA Removal and cDNA Synthesis SuperMix Kit. Real-time RT-PCR mixtures were composed of 10 μL of 2× TransStart ® Tip Green Qpcr SuperMix, 1 μL of cDNA, 4-pmol concentration of each primer, and nuclease-free water to a final volume of 20 μL. The OsACTIN gene served as an internal control for quantitative RT-PCR (Table S2). Primer pairs for defence-related genes were used to detect the gene expression levels (Table S2). All results were normalized by the OsACTIN expression, and relative changes in gene expression level were calculated by the comparative Ct method (Applied Biosystem, Foster City, CA). Amplification conditions were set as follows: 30 s at 94°C, then 45 cycles consisting of 5 s at 94°C and 30 s at 60°C. For each gene, data were taken from three independent biological replicates.
Statistical analysis
For each treatment, mean value served as a replicate and the experiment was repeated three times. Data from three replicates was subjected to analysis of variance (ANOVA) using DPS 5.0 and means were separated using Least Significant Difference (LSD) test. Means followed by the same letter in a column are not significantly different (P < 0.05) according to the LSD test.
Results
Identification of strain BJ-1
The strain BJ-1 was isolated from a contaminated M. oryzae culture plate, and showed strong inhibition to mycelial growth of M. oryzae. After incubation for 24 h on LB plates at 37°C, colonies were flavescent, glossy and had a strong odour. BJ-1 was rod-shaped with a Gram-positive staining reaction. Furthermore, BJ-1 produced short clavate endospores. Phenotypic microarray analyses conducted using the Biolog GEN III microplate system revealed that strain BJ-1 can utilize 25 different carbon sources, and showed resistance against 37 chemicals (Table S3). The Biolog system selected B. subtilis subsp. spizizenii as the highest match with a SIM of 0.711 based on the Microlog Gram-positive database. Successful PCR reactions with 16S primers 27f and 1492r resulted in single bands of ~1.5 kb observed on a 1% agarose gel. Fragments were sent for sequencing, and comparison with sequences in GenBank showed 100% similarity with B. subtilis (accession no. HM055602.1). Phylogenetic analyses based on 16S rDNA sequences of 16 sequences using the Neighbour-Joining method with a tree rooted on B. amyloliquefaciens showed that strain BJ-1 was most similar to B. subtilis (). Hence, the strain BJ-1 was identified as Bacillus subtilis based on morphological observations, Gram staining, Biolog identification and 16S rDNA sequence analysis.
Antagonism of BJ-1 to various plant pathogenic fungi
The dual culture test showed that B. subtilis strain BJ-1 inhibited mycelial growth of various plant pathogens on PDA (). Among 12 tested fungi, the inhibition zones ranged from 2.7 to 25.7 mm wide, with the most inhibitory effects (12.3–25.7 mm) against Ustilaginoidea virens, M. oryzae, Phoma aguilegiicola, Alternaria alternata, Botrytis cinerea, C. higginsianum and Stagonosporopsis cucurbitacearum, while less inhibitory activity (2.7–8.3 mm) was found against Sclerotinia sclerotiorum, Botryosphaeria dothidea, Helicobasidium mompa, Ceratobasidium sp. and Fusarium oxysporum.
Identification and expression of antifungal genes from BJ-1
To determine the potential antimicrobial compounds produced by BJ-1, 12 primer pairs were used in PCR amplification to detect antimicrobial biosynthetic genes in the genomic DNA (Table S1). Lipopeptide antibiotics produced by B. subtilis include the surfactin, iturin and fengycin families (Kim et al., Citation2009). The results of PCR screening revealed that three surfactin family antifungal genes (sfp, srfAA and SPAB-ERIB), two fengycin family antifungal genes (fenB and FENCEA), two subtilin antifungal genes (spa and spaS) and two iturin family bacilysin antifungal genes (BAC and BACD) were present in BJ-1, but three other iturin family genes (ituA, ituC and ituD) were not detected ().
Expression levels of nine detected antimicrobial biosynthetic genes were further evaluated using RT-PCR, and the results showed that seven genes – srfAA, SPAB-ERIB, fenB, FENCEA, spa, spaS and BACD – could be expressed in BJ-1 after incubation in LB and especially SPAB-ERIB, spaS and FENCEA were highly expressed ().
Suppression of mycelial growth of M. oryzae by culture filtrates of BJ-1
In the control, dry weight of 7-day cultures of M. oryzae was 6.5 mg/mL, whereas dry weight was significantly (P < 0.05) decreased from 2.8 to 0.3 mg mL−1 after treatment with different concentrations of culture filtrates from 0.5 to 20%, and mycelial growth was significantly inhibited by 56.3 to 94.7%, respectively (). Moreover, hyphae of M. oryzae treated with 0.5% BJ-1 culture filtrate were degraded and lysed while the control showed normal growth (). These results confirm that culture filtrates of B. subtilis BJ-1 suppress mycelial growth of M. oryzae.
Table 2. Suppression of mycelial growth of M. oryzae in vitro using culture filtrates of B. subtilis BJ-1.
Effects of culture filtrates of BJ-1 on conidial germination and appressorial formation of M. oryzae in vitro
Following incubation at 28°C for 18 h in darkness, conidial germination and appressorial formation were 98.7% and 94.0% in the control (), respectively, while culture filtrates did not affect germination of conidia but induced abnormally swollen germ tubes and decreased appressorial formation rates (). When concentrations of culture filtrates were increased from 0.5% to 20%, rates of germ tube malformation increased from 96.4% to 100%, and appressorial formation rates decreased from 47.3% to 0.3% (). The results showed that culture filtrates of B. subtilis BJ-1 could cause significant malformation of germ tubes and inhibit appressorial formation of M. oryzae.
Table 3. Effects of B. subtilis BJ-1 on conidial germination and appressorial formation of M. oryzae on plastic coverslips.
Effect of BJ-1 on rice blast development on detached rice leaves
After inoculation and incubation at 28°C for 3 d, typical blast disease spots were observed and mean lesion area was 56.4 mm2 among inoculated control plants (, ). In treatments with 2.5% culture broth, significantly (P = 0.05) reduced symptoms were observed, with lesion areas averaging only 1.4 mm2, while no symptoms were found with the 5% or 10% culture broth treatments (, ). Similarly, when conidial suspensions were mixed with culture filtrates at 5% or 10%, no lesions were observed, and even at 2.5% culture filtrate, lesions were smaller at 3.3 mm2 compared with the control at 54.1 mm2 (, ). Furthermore, washed bacterial cells of BJ-1 at 1 × 108 CFU mL−1 in conidial suspensions strongly inhibited rice blast development on detached rice leaves, while 1 × 107 CFU mL−1 bacterial cells only slightly reduced lesion expansion compared with the control (, ). Hence, culture broth, culture filtrates and bacterial cells of B. subtills BJ-1 all significantly suppressed M. oryzae development on rice leaves.
Table 4. Control efficacy of culture broth and culture filtrate of B. subtilis BJ-1 on rice blast development on detached rice leaves.
Table 5. Control efficacy of bacterial suspension of B. subtilis BJ-1 on rice blast development on detached rice leaves.
Effect of BJ-1 on rice blast development in pot experiments and expression of defence-related genes in rice plants
The culture broth of BJ-1 attained effective control of rice blast in the greenhouse by two approaches, spraying and seed treatments (, ). The disease index of plants inoculated with M. oryzae was 63.1 after 6 d at 28°C, and the leaves were shrivelled (, ). However, when sprayed with 10% culture broth and conidial suspensions simultaneously, the disease index decreased to 30.7 and only small lesions were observed (, ). After spraying with the fungicide tricyclazole, disease index was 16.6 and the control efficacy was 73.0% (, ). More importantly, when rice seeds were soaked with 3-day-old culture broth for 24 h and then planted, the disease index was only 15.8 and the control efficacy reached 74.3% (, ), not significantly different from tricyclazole treatment. Moreover, compared with the control at 28.5 cm, plant heights increased significantly after seed treatment with BJ-1 culture broth to 41.4 cm (, ).
Table 6. Control efficacy of culture broth of B. subtilis BJ-1 against rice blast in the greenhouse.
The expression patterns of two PR genes and four genes known to function in salicylate (SA)- or jasmonate (JA)-dependent defence signalling pathways in rice plants were examined after seed treatment with culture broth of BJ-1. Among the six defence-related genes, expression levels of five genes – OsPR1a (acidic PR protein), OsPR10 (ribonuclease), OsPAL (phenylalanine ammonia-lyase), OsICS1 (isochorismate synthase 1) and OsAOS2 (allene oxide synthase 2) were significantly induced ().
Discussion
Numerous B. subtilis strains have been described that can successfully control plant diseases (Cawoy et al., Citation2011). In this study, strain BJ-1 was isolated from a contaminated M. oryzae culture plate and identified as B. subtilis based on morphology, Biolog GEN III MicroPlate™ test and 16S rDNA sequence analysis. Strain BJ-1 strongly inhibited various plant pathogenic fungi in the plate assay, indicating that it possessed broad anti-fungal activity. Microscopic observations of M. oryzae hyphae after treatment with culture filtrates of BJ-1 showed abnormal swelling, lysis and complete degradation of the hyphal tip compared with the control (He et al., unpublished results). These results are similar to descriptions of Yang et al. (Citation2009) who reported that the antifungal compounds of B. subtilis may disrupt the hyphal cell wall. Gu et al. (Citation2017) also found that bacillomycin D produced by B. amyloliquefaciens FZB42 caused morphological changes in plasma membranes and cell walls of hyphae and conidia of F. graminearum. In our work, when conidia of M. oryzae were treated with culture filtrates of BJ-1, it led to malformation of germ tubes and reduction of appressorial formation. We deduce that the bioactive compounds from BJ-1 not only destroy cell walls of M. oryzae but also affect permeability of plasma membrane of fungal cells. This has been reported previously by Sha et al. (Citation2016).
The inhibitory activity of bioactive substances produced by B. subtilis BJ-1 appeared to be unchanged even after heating to 100°C for 30 min, and furthermore, the antimicrobial activity showed no drastic reduction within a wide range of pH values (2.0–12.0) (He et al., unpublished observations). Other studies have identified various bioactive compounds including small lipopeptides produced by other B. subtilis strains, in which the inhibitory effect was retained over a wide range of temperatures and pH values (Choi et al., Citation2012; Ramachandran et al., Citation2014).
The majority of Bacillus species have the capacity to produce two to four antimicrobial peptides (AMPs) in the culture filtrates (Mora et al., Citation2011; Yang et al., Citation2015). Lipopeptide antibiotics including the families surfactin, iturin and fengycin produced by B. subtilis are known to have strong antagonistic activity and stability (Kim et al., Citation2010; Pathak & Keharia, Citation2014). The surfactin family is one of the strongest biosurfactants to date, forming a biofilm layer on roots of plants but without direct antifungal capacity (Mihalache et al., Citation2017). The fengycin family (plipastatin and fengycin) and iturin family (iturin, mycosubtilin and bacillomycin) can disrupt surface tension of fungal cell membranes, leading to formation of micropores and leakage of important ions, finally causing cell death (Ongena & Jacques, Citation2008; Mihalache et al., Citation2017). It was reported that bacillomycin D produced by B. subtilis AU195 was an inhibitor of aflatoxin produced by Aspergillus flavus (Moyne et al., Citation2004). Earlier work reported that a B. subtilis mutant defective in surfactin production failed to protect Arabidopsis from infection by P. syringae (Bais et al., Citation2004). Fan et al. (Citation2017) reported that fengycin plays a major role in B. subtilis activity against apple ring rot caused by Botryosphaeria dothidea. In this study, B. subtilis BJ-1 showed significant antagonistic effects on growth of M. oryzae in vitro and in vivo. PCR and RT-PCR amplification showed the presence and expression of seven lipopeptide synthesis genes. However, three other iturin family antifungal genes (ituA, ituC and ituD) were not detected in genomic DNA of BJ-1, and associated synthesis genes may be lacking to synthesize these three lipopeptides.
In addition to direct antagonism, numerous studies reported that the induction of systemic host resistance also contributes to the biocontrol activities of B. subtilis (Choudhary & Johri, Citation2009; Lahlali et al., Citation2013). It was demonstrated that surfactin produced by B. subtilis S499 induced systemic resistance (Ongena et al., Citation2007). In experiments of rice seed treatments with culture broth of B. subtilis BJ-1, the results showed that this significantly suppressed disease and induced systemic resistance by potentially regulating defence-related genes in SA- and JA-dependent signalling pathways. Moreover, rice plant height after seed treatment was significantly higher compared with non-treated seeds, with similarities to enhanced plant growth after B. amyloliquefaciens FZB45 treatment as reported by Makarewicz et al. (Citation2006). Hence, we deduced that rice seeds treated with culture broth of BJ-1 might cause over-expression of related phytohormone genes to promote the growth of rice plants. Thus, B. subtilis BJ-1 showed diverse antibiotic capacities in the number and type of AMP genes, as well as stability of antibiotic compounds. In future studies, the major bioactive lipopeptides from B. subtilis BJ-1 for control of rice blast and other fungal plant disease should be identified.
Supplemental tables 1-3
Download MS Word (115.5 KB)Supplementary material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2018.1564792.
Additional information
Funding
References
- Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134:307–319.
- Caamaño-Antelo S, Fernández-No IC, Böhme K, Ezzat-Alnakip M, Quintela-Baluja M, Barros-Velázquez J, Calo-Mata P. 2015. Genetic discrimination of foodborne pathogenic and spoilage Bacillus spp. based on three housekeeping genes. Food Microbiol. 46:288–298.
- Cawoy H, Bettiol W, Fickers P, Ongena M. 2011. Bacillus-based biological control of plant diseases. In: Stoytcheva M, editor. Pesticides in the modern world-pesticides use and management. Croatia: InTech. 273–302
- Choi YS, Cho, Simkhada JR, Yoo JC. 2012. A novel thermotolerant and acidotolerant peptide produced by a Bacillus strain newly isolated from a fermented food (kimchi) shows activity against multidrug-resistant bacteria. Int J Antimicrob Agents. 40:80–83.
- Choudhary DK, Johri BN. 2009. Interactions of Bacillus spp. and plants with special reference to induced systemic resistance (ISR). Microbiol Res. 164:493–513.
- Dagdas YF, Yoshino K, Dagdas G, Ryder LS, Bielska E, Steinberg G, Talbot NJ. 2012. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science. 336:1590–1595.
- Fan HY, Ru JJ, Zhang YY, Wang Q, Li Y. 2017. Fengycin produced by Bacillus subtilis 9407 plays a major role in the biocontrol of apple ring rot disease. Microbiol Res. 199:89–97.
- Garcia-Gutiérrez L, Zeriouh H, Romero D, Cubero J, de Vicente A, Pérez-Garcia A. 2013. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence response. Microb Biotechnol. 6:264–274.
- Gu Q, Yang Y, Yuan QM, Shi GM, Wu LM, Lou ZY, Huo R, Wu HJ, Borriss R, Gao XW. 2017. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant pathogenic fungus Fusarium graminearum. Appl Environ Microb. 83. doi:10.1128/AEM.01075-17
- Hirooka T, Ishii H. 2013. Chemical control of plant diseases. J Gen Plant Pathol. 79:390–401.
- Kim GH, Lim MT, Hur JS, Yum KJ, Koh YJ. 2009. Biological control of tea anthracnose using an antagonistic bacterium of Bacillus subtilis isolated from tea leaves. Plant Pathol. 25:99–102.
- Kim PI, Ryu J, Kim YH, Chi YT. 2010. Production of biosurfactant lipopeptides Iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J Microbiol Biotechnol. 20:138–145.
- Kloepper JW, Ryu CM, Zhang S. 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology. 94:1259–1266.
- Lahlali R, Peng G, Gossen BD, Mcgregor L, Yu FQ, Hynes RK. 2013. Evidence that the biofungicide Serenade (Bacillus subtilis) suppresses clubroot on canola via antibiosis and induced host resistance. Phytopathology. 103:245–254.
- Leelasuphakul W, Sivanunsakul P, Phongpaichit S. 2006. Purification, characterization and synergistic activity of a β-1,3-glucanase and antibiotic extract from an antagonistic Bacillus subtilis NSRS 89-24 against rice blast and sheath blight. Enzyme Microb Technol. 38:990–997.
- Li GQ, Huang HC, Miao HJ, Erickson RS, Jiang DH, Xiao YN. 2006. Biological control of sclerotinia diseases of rapeseed by aerial applications of the mycoparasite Coniothyrium minitans. Eur J Plant Pathol. 114:345–355.
- Li QL, Jiang YH, Ning P, Zheng L, Huang JB, Li GQ, Jiang DH, Hsiang T. 2011. Suppression of Magnaporthe oryzae by culture filtrates of Streptomyces globisporus JK-1. Biol Control. 58:139–148.
- Makarewicz O, Dubrac S, Msadek T, Borriss R. 2006. Dual role of the PhoP∼P response regulator: bacillus amyloliquefaciens FZB45 phytase gene transcription is directed by positive and negative interaction with the phy C promoter. J Bacteriol. 188:6953–6965.
- Mihalache G, Balaes T, Gostin I, Stefan M. 2017. Lipopeptides produced by Bacillus subtilis as new biocontrol products against fusariosis in ornamental plants. Environ Sci Pollut R. doi:10.1007/s11356-017-9162-7
- Mora I, Cabrefiga J, Montesinos E. 2011. Antimicrobial peptide genes in Bacillus strains from plant environments. Int Microbiol. 14:213–223.
- Moyne AL, Cleveland TE, Tuzun S. 2004. Molecular characterization and analysis of the operon encoding the antifungal lipopeptide bacillomycin D. FEMS Microbiol Lett. 234:43–49.
- Ongena M, Jacques P. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16:115–125.
- Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P. 2007. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol. 9:1084–1090.
- Pathak KV, Keharia H. 2014. Identification of surfactins and iturins produced by potent fungal antagonist, Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) tree using mass spectrometry. J Biotechnol. 4:283–295.
- Pedersen EA, Reddy MS, Chakravarty P. 2010. Effect of three species of bacteria on damping-off, root rot development, and ectomycorrhizal colonization of lodgepole pine and white spruce seedlings. Forest Pathol. 29:123–134.
- Prabavathy VR, Mathivanan N, Murugesan K. 2006. Control of blast and sheath blight diseases of rice using antifungal metabolites produced by Streptomyces sp. PM5. Biol Control. 39:313–319.
- Ramachandran R, Chalasani AG, Lal R, Roy U. 2014. A broad-spectrum antimicrobial activity of Bacillus subtilis RLID 12.1. Sci World J. 2014:968487.
- Reynolds J, Moyes R, Breakwell DP. 2009. Differential staining of bacteria: endospore stain. Curr Protoc Microbiol. Appendix 3:3J.
- Sajitha KL, Dev SA. 2016. Quantification of antifungal lipopeptide gene expression levels in Bacillus subtilis B1 during antagonism against sapstain fungus on rubberwood. Biol Control. 96:78–85.
- Sha YX, Wang Q, Li Y. 2016. Suppression of Magnaporthe oryzae and interaction between Bacillus subtilis and rice plants in the control of rice blast. SpringerPlus. 5:1238.
- Thompson JD, Gibson T, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Unit 2. 3:1–22.
- Turner JT, Backman PA. 1991. Factors relating to peanut yield increases after seed treatment with Bacillus subtilis. Plant Dis. 75:347–353.
- US Environmental Protection Agency. 1997. Final risk assessment of Bacillus subtilis. Biotechnology publications. www.epa.gov/oppt/biotech/pubs/pdf/fra009.pdf.
- Yang D, Wang B, Wang J, Chen Y, Zhou M. 2009. Activity and efficacy of Bacillus subtilis strain NJ-18 against rice sheath blight and Sclerotinia stem rot of rape. Biol Control. 51:61–65.
- Yang LR, Quan X, Xue BG, Goodwin PH, Lu SB, Wang JH, Wei D. 2015. Isolation and identification of Bacillus subtilis strain YB-05 and its antifungal substances showing antagonism against Gaeumannomyces graminis var. tritici. Biol Control. 85:52–58.
- Zohora US, Ano T, Rahman MS. 2016. Biocontrol of Rhizoctonia solani K1 by iturin a producer Bacillus subtilis RB14 seed treatment in tomato plants. Adv Microbiol. 6:424–431.
